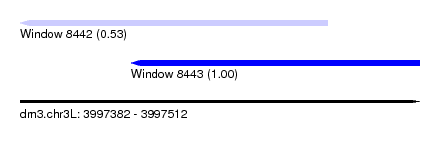
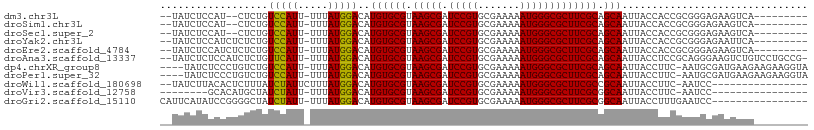
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,997,382 – 3,997,512 |
| Length | 130 |
| Max. P | 0.996325 |

| Location | 3,997,382 – 3,997,482 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.99 |
| Shannon entropy | 0.63764 |
| G+C content | 0.51814 |
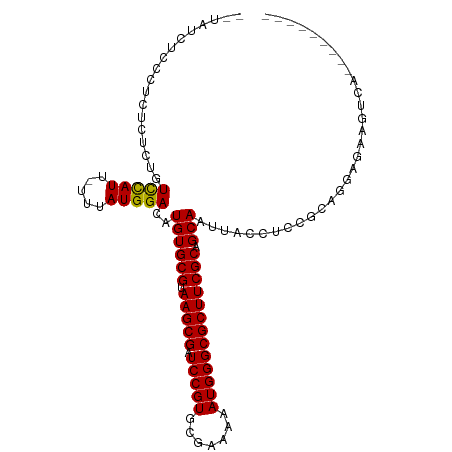
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.10 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3997382 100 - 24543557 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAA---------GUCAGAGCCGACCAGGC-CAAAUAGAUUCGC--AGGCCGAUAA ((((((.(((((.(((((.......)))))))))))))).)).......(((((((.(..---------..).(.(((.....)))-).......)))))--.))....... ( -27.60, z-score = 0.67, R) >droSim1.chr3L 3540930 100 - 22553184 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAA---------GUCAGAGCCGACCAGGC-CAAAUAGAUUCGC--AGGCCGAUAA ((((((.(((((.(((((.......)))))))))))))).)).......(((((((.(..---------..).(.(((.....)))-).......)))))--.))....... ( -27.60, z-score = 0.67, R) >droSec1.super_2 3989935 100 - 7591821 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAA---------GUCAGAGCCGACCAGGU-CAAAUAGAUUCGC--AGGCCGAUAA ((((((.(((((.(((((.......)))))))))))))).))....((.....)).....---------(((.(.(((...(..((-(.....)))..).--.))))))).. ( -26.90, z-score = 0.76, R) >droYak2.chr3L 4561732 100 - 24197627 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAA---------UUCAGAACCGACCAGGC-CAAAUAGAUUCGC--AGGCCGAUAA ((((((.(((((.(((((.......)))))))))))))).))..........(((.....---------.......)))....(((-(............--.))))..... ( -27.22, z-score = 0.36, R) >droEre2.scaffold_4784 6699103 100 - 25762168 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAA---------GUCAGAAACGACCAGGC-CAAAUAGAUUCGC--AGGCCGAUAA ((((((.(((((.(((((.......)))))))))))))).))....((.....)).....---------(((......)))..(((-(............--.))))..... ( -27.22, z-score = 0.34, R) >droAna3.scaffold_13337 1844456 111 - 23293914 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUCCGCAGGGAAGUCUGUC-CUGCCGGGCCGGACCCGGUACAAAUAGAUUCCCCAUGGCCGAUAA ((((((.(((((.(((((.......)))))))))))))).))......((.((.(((.(((((((.-.(((((((.....)))))))...))))))).)))...)).))... ( -43.90, z-score = -2.02, R) >dp4.chrXR_group8 645966 109 - 9212921 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUUC-AAUGCGAUGAAGAAGAAGGUAGCUCCAACCGAA--UAUGUAGAUGCCACACUGGCGAUAA ((((((.(((((.(((((.......)))))))))))))).)).((((((((-....(......)..))))))))...........--........((((.....)))).... ( -30.80, z-score = -0.78, R) >droPer1.super_32 153808 109 + 992631 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUUC-AAUGCGAUGAAGAAGAAGGUAGCUCCAACCGAA--UAUGUAGAUGCCACACUGGCGAUAA ((((((.(((((.(((((.......)))))))))))))).)).((((((((-....(......)..))))))))...........--........((((.....)))).... ( -30.80, z-score = -0.78, R) >droWil1.scaffold_180698 8766572 91 - 11422946 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCCGCAAUUACCUUC-AAUCCGAAACAGCUUGAAAUAGAUUUCACUUUA--GUGAUAA------------------ ((((((.(((((.(((((.......)))))))))))).))))......(((-((..(......).))))).......((((....--))))...------------------ ( -21.40, z-score = -0.49, R) >droVir3.scaffold_12758 5256 90 + 972128 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCGGCAAUUACCUUC-AAUCCAAAACAGCU-CCAACAGAUUCAUCAGUU--CCGAUAA------------------ ((((((.(((((.(((((.......)))))))))))))((......))...-........)))...-...........(((....--..)))..------------------ ( -17.80, z-score = 0.61, R) >droMoj3.scaffold_6680 22746199 82 + 24764193 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCGGCAAUUACCUUUGGCUCCAA----------GGGAGAUUCAUCCAUU--GCGAUAA------------------ ..((((.(((((.(((((.......))))))))))))))(((((..((((((....)))----------))).((....)).)))--)).....------------------ ( -27.40, z-score = -1.10, R) >droGri2.scaffold_15110 545794 92 - 24565398 UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCGGCAAUUACCUUUGAAUCCGAAACAGCUAGAAACAGAUUCAUCCGCU--GCGAUAA------------------ ........((((.(((((.......)))))))))(((((((.......((((....)))).......(((.....)))....)))--))))...------------------ ( -22.20, z-score = 0.61, R) >consensus UGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUCCGCAGGCGAA_________GUCAGAUCCGACCAGG__CAAAUAGAUUCGC__AGGCCGAUAA ((((((.(((((.(((((.......)))))))))))))).))...................................................................... (-17.52 = -17.44 + -0.08)
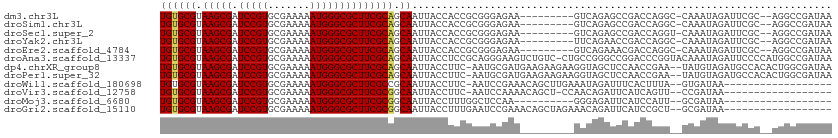
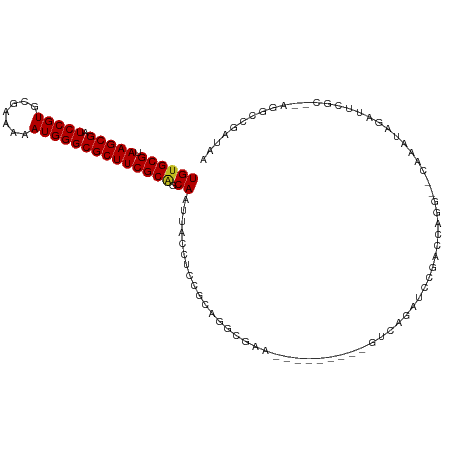
| Location | 3,997,418 – 3,997,512 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.81 |
| Shannon entropy | 0.45045 |
| G+C content | 0.48246 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.24 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3997418 94 - 24543557 --UAUCUCCAU--CUCUGUCCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAAGUCA--------- --..(((((..--...(((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).))............))))).....--------- ( -34.70, z-score = -3.19, R) >droSim1.chr3L 3540966 94 - 22553184 --UAUCUCCAU--CUCUGUCCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAAGUCA--------- --..(((((..--...(((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).))............))))).....--------- ( -34.70, z-score = -3.19, R) >droSec1.super_2 3989971 94 - 7591821 --UAUCUCCAU--CUCUGUCCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAAGUCA--------- --..(((((..--...(((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).))............))))).....--------- ( -34.70, z-score = -3.19, R) >droYak2.chr3L 4561768 96 - 24197627 --UAUCUCCAUCUCUCUGUCCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAAUUCA--------- --........(((((((((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).))...........)))))).....--------- ( -34.80, z-score = -3.42, R) >droEre2.scaffold_4784 6699139 96 - 25762168 --UAUCUCCAUCUCUCUGUCCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCACCGCGGGAGAAGUCA--------- --........(((((((((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).))...........)))))).....--------- ( -34.80, z-score = -3.17, R) >droAna3.scaffold_13337 1844495 104 - 23293914 --UAUCUCUCCAUCUCUGUUCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUCCGCAGGGAAGUCUGUCCUGCCG- --..............(((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).)).........((((((.......))))))..- ( -31.80, z-score = -1.81, R) >dp4.chrXR_group8 646003 102 - 9212921 ----UAUCUCCCUGUCUGUCCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUUC-AAUGCGAUGAAGAAGAAGGUA ----............(((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).))..(((((((-....(......)..))))))) ( -33.60, z-score = -2.73, R) >droPer1.super_32 153845 102 + 992631 ----UAUCUCCCUGUCUGUCCAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUUC-AAUGCGAUGAAGAAGAAGGUA ----............(((((((.-...)))))))((((((.(((((.(((((.......)))))))))))))).))..(((((((-....(......)..))))))) ( -33.60, z-score = -2.73, R) >droWil1.scaffold_180698 8766607 89 - 11422946 --UAUCUUACACUCUUUAUCUAUUCUUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCCGCAAUUACCUUC-AAUCC---------------- --................(((((.....)))))..((((((.(((((.(((((.......)))))))))))).)))).........-.....---------------- ( -19.90, z-score = -1.49, R) >droVir3.scaffold_12758 5290 82 + 972128 --------GCACAUGCUAUCUAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCGGCAAUUACCUUC-AAUCC---------------- --------(((((((((((.....-...)))).)))))))(((((((.(((((.......)))))))))).))((......))...-.....---------------- ( -25.70, z-score = -2.11, R) >droGri2.scaffold_15110 545829 91 - 24565398 CAUUCAUAUCCGGGGCUAUCUAUU-UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCGGCAAUUACCUUUGAAUCC---------------- .(((((.....((.(((.(((((.-...))))).....(((.(((((.(((((.......)))))))))))))))).....))..)))))..---------------- ( -24.40, z-score = -0.51, R) >consensus __UAUCUCCCUCUCUCUGUCCAUU_UUUAUGGACAUGUGCGUAAGCGAUCCGUGCGAAAAAUGGGCGCUUCGCAGCAAUUACCUCCGCAGGAGAAGUCA_________ ..................(((((.....)))))..((((((.(((((.(((((.......))))))))))))).)))............................... (-21.52 = -21.24 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:57 2011