
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,988,054 – 3,988,167 |
| Length | 113 |
| Max. P | 0.996135 |

| Location | 3,988,054 – 3,988,167 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Shannon entropy | 0.36027 |
| G+C content | 0.49871 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -20.98 |
| Energy contribution | -23.18 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
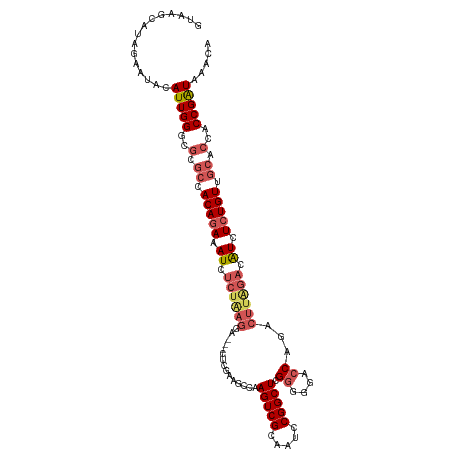
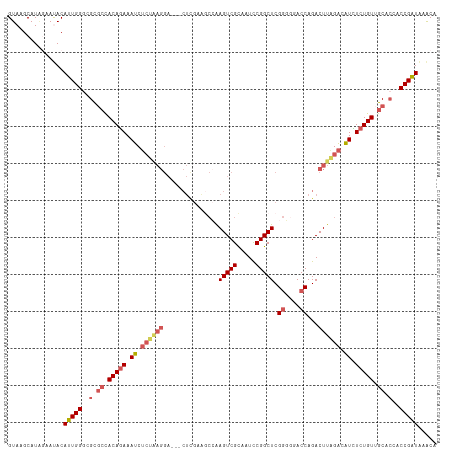
>dm3.chr3L 3988054 113 + 24543557 GUAAGCAUAGAAUACAUUGGGCGCGCUACAGAAAUCUCUAAGGA---CUUGAAGCCAAGUCGCAAUCCGGCUCGGGUGACCAGACUUAGACAUCUCUGUUGCACAACCGAUAAACA ...............(((((..(.((.(((((.((.((((((((---((((....))))))((......))..((....))...)))))).)).))))).)).)..)))))..... ( -35.60, z-score = -2.93, R) >droSim1.chr3L 3531670 113 + 22553184 GUAAGCAUAGAAUACAUUGGGCGCGCCACAGAAAUCUCUGAGGA---CUCGAAGCCAAGUCGCAAUUCGGCUCGGGGGACCAGACUUAGAAAUCUCUGUUGCACUACCGAUAAACA ...............(((((..(.((.(((((.((.((((((((---..((((((......))..))))..))((....))...)))))).)).))))).)).)..)))))..... ( -32.90, z-score = -1.59, R) >droSec1.super_2 3980688 113 + 7591821 GUAAGCAUAGAAUACAUUGGGCGCGCCACAGAAAUCUCUAAGGA---CUCGAAGCCAAGUCGCAAUCCGGCUCGGGGGACCAGACUUAGACAUCUCUGUGGCACUACCGAUAAACA ...............(((((..(.((((((((.((.((((((..---...((.(((............)))))((....))...)))))).)).)))))))).)..)))))..... ( -38.20, z-score = -3.11, R) >droYak2.chr3L 4552172 96 + 24197627 GUAAGCAUCAAACACAUUGGGCGCUACACAAAAAUC--------------------AAGUCGUGAUCCGGCUCGUUGGACCAGACUUGGGGAUUUCUGUUGCAACGCCGAUGAACA ..............(((((((.((.(((.(((..((--------------------(((((.((.(((((....))))).)))))))))...))).))).))..).)))))).... ( -26.10, z-score = -0.68, R) >droEre2.scaffold_4784 6689679 116 + 25762168 GUAAGCAUGAAACACAUUGGCCGUGCCACAGAAAUCUCUGAGGAAAUCUUGAGACGGAGUCGUAAUCCGGCUCGGUGGACCAGACUUGGACGUUUCUGUUGCACCACCGGUAAGCA ...............(((((..((((.((((((((.((..((........(((.((((.......)))).)))((....))...))..)).)))))))).))))..)))))..... ( -38.60, z-score = -1.41, R) >consensus GUAAGCAUAGAAUACAUUGGGCGCGCCACAGAAAUCUCUAAGGA___CUCGAAGCCAAGUCGCAAUCCGGCUCGGGGGACCAGACUUAGACAUCUCUGUUGCACCACCGAUAAACA ...............(((((..(.((.(((((.((.((((((...............(((((.....))))).((....))...)))))).)).))))).)).)..)))))..... (-20.98 = -23.18 + 2.20)

| Location | 3,988,054 – 3,988,167 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Shannon entropy | 0.36027 |
| G+C content | 0.49871 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -22.96 |
| Energy contribution | -24.16 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
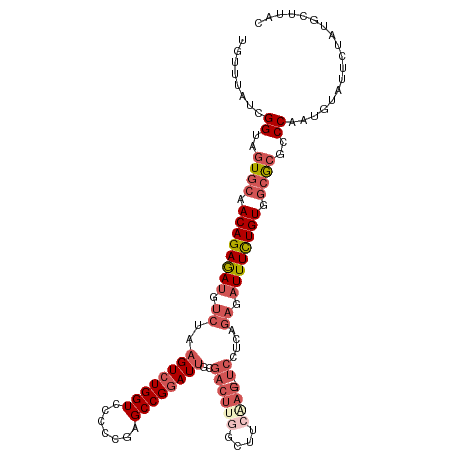
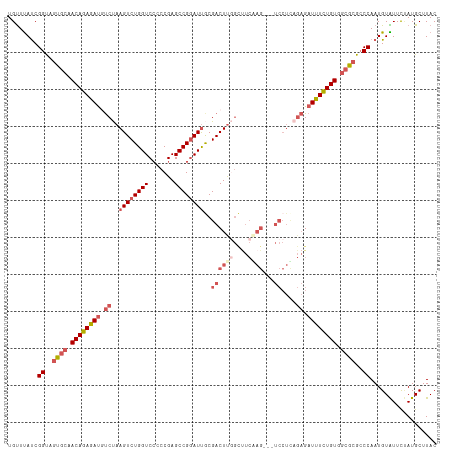
>dm3.chr3L 3988054 113 - 24543557 UGUUUAUCGGUUGUGCAACAGAGAUGUCUAAGUCUGGUCACCCGAGCCGGAUUGCGACUUGGCUUCAAG---UCCUUAGAGAUUUCUGUAGCGCGCCCAAUGUAUUCUAUGCUUAC ........((.(((((.((((((((.((((((((((((.......))))))....((((((....))))---)))))))).)))))))).))))).)).................. ( -41.80, z-score = -3.97, R) >droSim1.chr3L 3531670 113 - 22553184 UGUUUAUCGGUAGUGCAACAGAGAUUUCUAAGUCUGGUCCCCCGAGCCGAAUUGCGACUUGGCUUCGAG---UCCUCAGAGAUUUCUGUGGCGCGCCCAAUGUAUUCUAUGCUUAC ........((..((((.((((((((((((.((.((.(....(((((.((.....)).)))))...).))---..)).)))))))))))).))))..)).................. ( -35.10, z-score = -1.91, R) >droSec1.super_2 3980688 113 - 7591821 UGUUUAUCGGUAGUGCCACAGAGAUGUCUAAGUCUGGUCCCCCGAGCCGGAUUGCGACUUGGCUUCGAG---UCCUUAGAGAUUUCUGUGGCGCGCCCAAUGUAUUCUAUGCUUAC ........((..(((((((((((((.((((((((((((.......))))))....((((((....))))---)))))))).)))))))))))))..)).................. ( -47.50, z-score = -5.09, R) >droYak2.chr3L 4552172 96 - 24197627 UGUUCAUCGGCGUUGCAACAGAAAUCCCCAAGUCUGGUCCAACGAGCCGGAUCACGACUU--------------------GAUUUUUGUGUAGCGCCCAAUGUGUUUGAUGCUUAC ....(((((((((((((.((((((((...(((((((((((........)))))).)))))--------------------)))))))))))))))))(.....)...))))..... ( -35.70, z-score = -4.06, R) >droEre2.scaffold_4784 6689679 116 - 25762168 UGCUUACCGGUGGUGCAACAGAAACGUCCAAGUCUGGUCCACCGAGCCGGAUUACGACUCCGUCUCAAGAUUUCCUCAGAGAUUUCUGUGGCACGGCCAAUGUGUUUCAUGCUUAC .((.....(((.((((.((((((..(((..((((((((.......))))))))..)))...(((((.((.....))..))))))))))).)))).))).(((.....))))).... ( -36.40, z-score = -1.37, R) >consensus UGUUUAUCGGUAGUGCAACAGAGAUGUCUAAGUCUGGUCCCCCGAGCCGGAUUGCGACUUGGCUUCAAG___UCCUCAGAGAUUUCUGUGGCGCGCCCAAUGUAUUCUAUGCUUAC ........((..((((.((((((((.((..((((((((.......))))))))((......))...............)).)))))))).))))..)).................. (-22.96 = -24.16 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:56 2011