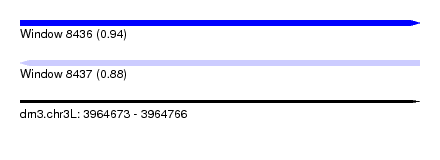
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,964,673 – 3,964,766 |
| Length | 93 |
| Max. P | 0.938865 |

| Location | 3,964,673 – 3,964,766 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 69.93 |
| Shannon entropy | 0.64250 |
| G+C content | 0.43797 |
| Mean single sequence MFE | -23.44 |
| Consensus MFE | -9.29 |
| Energy contribution | -9.49 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
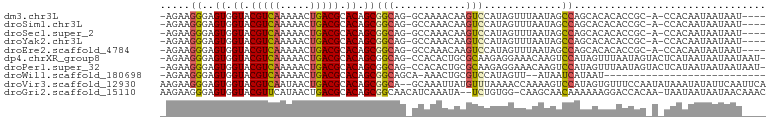
>dm3.chr3L 3964673 93 + 24543557 ----AUUAUUAUUGUGG-U-GCGGUGUGUGCUGGCUAUUAAACUAUGGACUUGUUUUGC-CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- ----.........((((-(-((((((...((.(((....((((.........)))).))-).))))))))(((((((...))))))))))))........- ( -27.00, z-score = -2.38, R) >droSim1.chr3L 3508385 93 + 22553184 ----AUUAUUAUUGUGG-U-GCGGUGUGUGCUGGCUAUUAAACUAUGGACUUGUUUGGC-CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- ----.........((((-(-((((((...((.(((((...(((.........)))))))-).))))))))(((((((...))))))))))))........- ( -27.60, z-score = -2.00, R) >droSec1.super_2 3957596 93 + 7591821 ----AUUAUUAUUGUGG-U-GCGGUGUGUGCUGGCUAUUAAACUAUGGACUUGUUUGGC-CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- ----.........((((-(-((((((...((.(((((...(((.........)))))))-).))))))))(((((((...))))))))))))........- ( -27.60, z-score = -2.00, R) >droYak2.chr3L 4527304 93 + 24197627 ----AUUAUUAUUGUGG-U-GCGGUGUGUGCUGGCUAUUAAACUAUGGACUUGUUUGGC-CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- ----.........((((-(-((((((...((.(((((...(((.........)))))))-).))))))))(((((((...))))))))))))........- ( -27.60, z-score = -2.00, R) >droEre2.scaffold_4784 6666272 93 + 25762168 ----AUUAUUAUUGUGG-U-GCGGUGUGUGCUGGCUAUUAAACUAUGGACUUGUUUGGC-CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- ----.........((((-(-((((((...((.(((((...(((.........)))))))-).))))))))(((((((...))))))))))))........- ( -27.60, z-score = -2.00, R) >dp4.chrXR_group8 610548 98 + 9212921 -AUUAUUAUUAUUAUGAGUACUAUUAAACUAUGGACUUGUUUCCUCUUGCGCAGUGUGG-CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- -............(..(((.((((......)))))))..)........((((((.....-))).))).(((((((((...)))))))))...........- ( -22.20, z-score = -1.66, R) >droPer1.super_32 118005 98 - 992631 -AUUAUUAUUAUUAUGAGUACUAUUAAACUAUGGACUUGUUUCCUCUUGCGCAGUGUGG-CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- -............(..(((.((((......)))))))..)........((((((.....-))).))).(((((((((...)))))))))...........- ( -22.20, z-score = -1.66, R) >droWil1.scaffold_180698 8726190 70 + 11422946 ---------------------------AUUAUGAUUAU--AACUAUGGACGCAGUUU-UGCUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU- ---------------------------...........--......(((.((((...-..))))....(((((((((...)))))))))...))).....- ( -17.30, z-score = -2.28, R) >droVir3.scaffold_12930 50487 99 - 74342 UGAAUUGAAUAUAUUUAUAUUGGAAACACUAUGGACUUUUGGUUUUAAACAUAAUUUGC--UGCCGCUGUGCGUCAGUUAUUGACGUACCACUCCCUUCUU .(((..((.....((((((.((....)).)))))).....(((..((((.....)))).--.)))...(((((((((...)))))))))...))..))).. ( -19.40, z-score = -1.46, R) >droGri2.scaffold_15110 505316 97 + 24565398 GUUUGUUAUUAUUAUUA-UUGUGGUCCUUUUUUGUUGCUUG-CCACAGA--UAUUUGAUGUUGCCGCUGUGCGUCAGUUAUGAACGUACCACUCCCUUCUU .................-..(((((.(...((..(.(((.(-((((((.--...............))))).)).))).)..)).).)))))......... ( -15.89, z-score = -0.51, R) >consensus ____AUUAUUAUUGUGG_U_GCGGUGUGUGCUGGCUAUUAAACUAUGGACUUAUUUGGC_CUGCCGCUGUGCGUCAGUUUUUGACGUACCACUCCCUUCU_ ....................................................................(((((((((...)))))))))............ ( -9.29 = -9.49 + 0.20)


| Location | 3,964,673 – 3,964,766 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 69.93 |
| Shannon entropy | 0.64250 |
| G+C content | 0.43797 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -8.11 |
| Energy contribution | -8.20 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
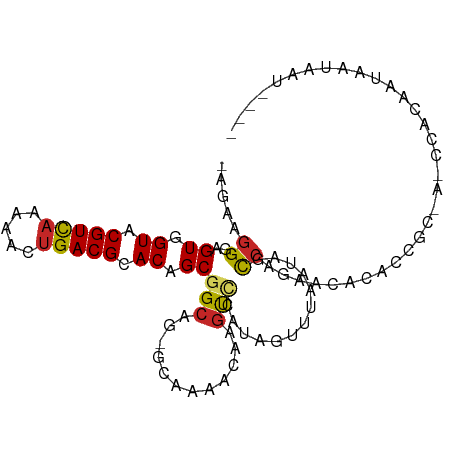
>dm3.chr3L 3964673 93 - 24543557 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG-GCAAAACAAGUCCAUAGUUUAAUAGCCAGCACACACCGC-A-CCACAAUAAUAAU---- -.....((.(((((.(((((.....)))))....(((((..-...((((.........))))....))).))....)))))-.-))...........---- ( -20.90, z-score = -1.89, R) >droSim1.chr3L 3508385 93 - 22553184 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG-GCCAAACAAGUCCAUAGUUUAAUAGCCAGCACACACCGC-A-CCACAAUAAUAAU---- -.....((.(((((.(((((.....)))))....(((((.(-(.(......).)).((......))))).))....)))))-.-))...........---- ( -21.20, z-score = -1.76, R) >droSec1.super_2 3957596 93 - 7591821 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG-GCCAAACAAGUCCAUAGUUUAAUAGCCAGCACACACCGC-A-CCACAAUAAUAAU---- -.....((.(((((.(((((.....)))))....(((((.(-(.(......).)).((......))))).))....)))))-.-))...........---- ( -21.20, z-score = -1.76, R) >droYak2.chr3L 4527304 93 - 24197627 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG-GCCAAACAAGUCCAUAGUUUAAUAGCCAGCACACACCGC-A-CCACAAUAAUAAU---- -.....((.(((((.(((((.....)))))....(((((.(-(.(......).)).((......))))).))....)))))-.-))...........---- ( -21.20, z-score = -1.76, R) >droEre2.scaffold_4784 6666272 93 - 25762168 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG-GCCAAACAAGUCCAUAGUUUAAUAGCCAGCACACACCGC-A-CCACAAUAAUAAU---- -.....((.(((((.(((((.....)))))....(((((.(-(.(......).)).((......))))).))....)))))-.-))...........---- ( -21.20, z-score = -1.76, R) >dp4.chrXR_group8 610548 98 - 9212921 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG-CCACACUGCGCAAGAGGAAACAAGUCCAUAGUUUAAUAGUACUCAUAAUAAUAAUAAU- -......(((((((.(((((.....))))).)).(((.(((-.....))))))....(((......)))............)))))..............- ( -21.20, z-score = -1.55, R) >droPer1.super_32 118005 98 + 992631 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG-CCACACUGCGCAAGAGGAAACAAGUCCAUAGUUUAAUAGUACUCAUAAUAAUAAUAAU- -......(((((((.(((((.....))))).)).(((.(((-.....))))))....(((......)))............)))))..............- ( -21.20, z-score = -1.55, R) >droWil1.scaffold_180698 8726190 70 - 11422946 -AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAGCA-AAACUGCGUCCAUAGUU--AUAAUCAUAAU--------------------------- -.....(((((.((.(((((.....))))).)).)).((((..-...)))).)))......--...........--------------------------- ( -16.50, z-score = -1.65, R) >droVir3.scaffold_12930 50487 99 + 74342 AAGAAGGGAGUGGUACGUCAAUAACUGACGCACAGCGGCA--GCAAAUUAUGUUUAAAACCAAAAGUCCAUAGUGUUUCCAAUAUAAAUAUAUUCAAUUCA ..(((..(((((((.(((((.....))))).))...((.(--(((...((((..(..........)..)))).)))).))..........)))))..))). ( -18.70, z-score = -1.35, R) >droGri2.scaffold_15110 505316 97 - 24565398 AAGAAGGGAGUGGUACGUUCAUAACUGACGCACAGCGGCAACAUCAAAUA--UCUGUGG-CAAGCAACAAAAAAGGACCACAA-UAAUAAUAAUAACAAAC .........(((((.((((.......)))((...((.(((..........--..))).)-)..)).........).)))))..-................. ( -13.40, z-score = -0.25, R) >consensus _AGAAGGGAGUGGUACGUCAAAAACUGACGCACAGCGGCAG_GCAAAACAAGUCCAUAGUUUAAUAGCCAGAACACACCGC_A_CCACAAUAAUAAU____ .....((..((.((.(((((.....))))).)).))(((............))).............))................................ ( -8.11 = -8.20 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:53 2011