| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,964,122 – 3,964,217 |
| Length | 95 |
| Max. P | 0.969159 |

| Location | 3,964,122 – 3,964,217 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 68.55 |
| Shannon entropy | 0.63400 |
| G+C content | 0.51070 |
| Mean single sequence MFE | -17.71 |
| Consensus MFE | -10.36 |
| Energy contribution | -10.36 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3964122 95 + 24543557 --CAGCGAGUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCUUAGCCUACCUCACUGUACCACCCCCUUUCUCCAA---CCCAUUUC-UCGCCCAUUUCGC--- --..(((((.....(((((.(((((.---...))))).))))).......(((......))).................---.......)-)))).........--- ( -16.40, z-score = -1.98, R) >dp4.chrXR_group8 609859 86 + 9212921 ----GCGAGUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCUUAGCCUACCUCACUGUCGUCUCCACCAGCACACC---CCUAUGCCGUACAC----------- ----....((((..(((((.(((((.---...))))).))))).............................(((....---....))).))))..----------- ( -16.00, z-score = -0.14, R) >droPer1.super_32 117293 86 - 992631 ----GCGAGUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCUUAGCCUACCUCACUGUCGUCUCCACCAGCACACC---CCUAUCCCGUACAC----------- ----....((((..(((((.(((((.---...))))).)))))..............(((..(....)..)))......---........))))..----------- ( -15.00, z-score = -0.60, R) >droGri2.scaffold_15110 504458 83 + 24565398 UGCGGCAAAUACAUGCAACAAAUAGUAGUCAACUGUUGGUUGCCUUUGCCUACCUCACUGUGCUUGCCACUCGCCACA----CUUGC-------------------- .((((((((.....(((((.((((((.....)))))).))))).))))))........((((...((.....))))))----...))-------------------- ( -19.90, z-score = -0.23, R) >droMoj3.scaffold_6680 22711656 94 - 24764193 --UGCGGCAUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCCUAGCCUACCUCACUGUGC---UCGAGCACCCA-----CCCUCACUCUCACACACACACUCGC --.(((((.((...(((((.(((((.---...))))).)))))..))))).........((((---....))))...-----.......................)) ( -19.40, z-score = -0.82, R) >droWil1.scaffold_180698 8725349 102 + 11422946 --UAGGGCAUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCUUAGCCUACCUCACUGAAUAUGUCGGGAACCCCAAUGCUUCCCUCUCUUUCUCACUGACUCUC --..((((.((...(((((.(((((.---...))))).)))))..))))))...(((.(((....(..(((((.(.....).)))))...)....))).)))..... ( -22.60, z-score = -1.13, R) >droSim1.chr3L 3507849 95 + 22553184 --CAGCGAGUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCUUAGCCUACCUCACUGUACCACCCCCUUUCUCCAA---CCCAUUUC-UCGCCCAUUUCGC--- --..(((((.....(((((.(((((.---...))))).))))).......(((......))).................---.......)-)))).........--- ( -16.40, z-score = -1.98, R) >droSec1.super_2 3957061 95 + 7591821 --CAGCGAGUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCUUAGCCUACCUCACUGUACCACCCCCUUUCUCCAA---CCCAUUUC-UCGCCCAUUUCGC--- --..(((((.....(((((.(((((.---...))))).))))).......(((......))).................---.......)-)))).........--- ( -16.40, z-score = -1.98, R) >droAna3.scaffold_13337 1807623 93 + 23293914 --UAGCGAGUACAUGCAACAAAUAGU---CAACUGUUGGUUGCCUUAGCCUACCUCACUGUACCACCCACCACCCACCU---CCCACCGAAUCUCGCACUC------ --..(((((.....(((((.(((((.---...))))).))))).......(((......))).................---..........)))))....------ ( -17.30, z-score = -2.04, R) >consensus __CAGCGAGUACAUGCAACAAAUAGU___CAACUGUUGGUUGCCUUAGCCUACCUCACUGUACCACCCACUAGCCCCAA___CCCAUUCC_UCGCCCA_U_______ ..............(((((.((((((.....)))))).)))))................................................................ (-10.36 = -10.36 + 0.00)

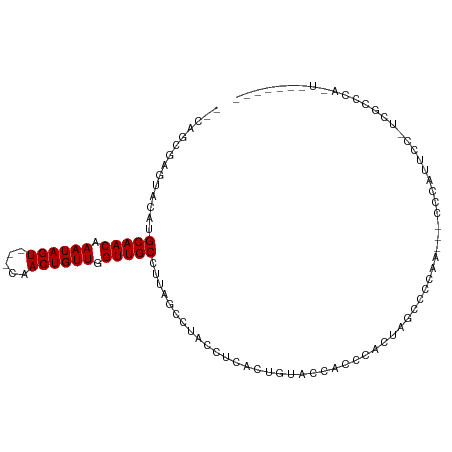
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:51 2011