
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,941,608 – 3,941,728 |
| Length | 120 |
| Max. P | 0.836821 |

| Location | 3,941,608 – 3,941,728 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.68401 |
| G+C content | 0.49139 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -16.01 |
| Energy contribution | -15.84 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.77 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3941608 120 + 24543557 AACCACAAUUGACGCACCUGAUUGUGGAUUGGCUUGAUCUUGCAGUUGGCCAGCAAGCGAGUCAACUGUUCCGAGUUGAAAUUGGAUACGCCGAUGCUCUUGGUCAGGCCCAGUUCCACG ..((((((((.........))))))))...(((((((((..(((((((((..(....)..)))))))))...((((....(((((.....)))))))))..))))))))).......... ( -42.10, z-score = -2.11, R) >droSim1.chr3L 3482301 120 + 22553184 AACCACAAUUGACGCACCUGAUUGUGGAUUGGCUUGAUCUUGCAGUUGGCCAGCAAGCGAGUCAACUGUUCCGAGUUGAAAUUGGAUACGCCGAUGCUCUUGGUCAGGCCCAGUUCCACG ..((((((((.........))))))))...(((((((((..(((((((((..(....)..)))))))))...((((....(((((.....)))))))))..))))))))).......... ( -42.10, z-score = -2.11, R) >droSec1.super_2 3934800 120 + 7591821 AACCACAAUUGACGCACCUGAUUGUGGAUUGGCUUGAUCUUGCAGUUGGCCAGCAAGCGAGUCAACUGUUCCGAGUUGAAAUUGGAUACGCCGAUGCUCUUGGUCAGGCCCAGUUCCACG ..((((((((.........))))))))...(((((((((..(((((((((..(....)..)))))))))...((((....(((((.....)))))))))..))))))))).......... ( -42.10, z-score = -2.11, R) >droYak2.chr3L 4502754 120 + 24197627 AACCACAAAUGACGUACCUGGUUGUGGAUGGGCUUGAUCUUGCAGUUGGCCAGCAGGCGAGUCAACUGCUCCGAGUUGAAAUUGGAUACGCCGAUGCUCUUGGUCAGGCCCAGUUCCACC (((((.............)))))((((((((((((((((..(((((((((..(....)..)))))))))...((((....(((((.....)))))))))..)))))))))))..))))). ( -49.22, z-score = -3.00, R) >droEre2.scaffold_4784 6642493 120 + 25762168 AACCACAAUUGACGCACCUGAUUGUGGAUUGGCUUGAUCUUGCAGUUGGCCAGCAAGCGAGUCAACUGCUCCGAGUUGAAAUUGGAUACGCCGAUGCUCUUGGUCAGGCCCAGUUCCACA ..((((((((.........))))))))...(((((((((..(((((((((..(....)..)))))))))...((((....(((((.....)))))))))..))))))))).......... ( -44.40, z-score = -2.54, R) >droAna3.scaffold_13337 13797415 120 - 23293914 AACCGCAAUUUACGUACCUGGUUGUGAAUGGGCUUGAUCUUGCAGUUGGCCAGAAGCCGAGUCAGUUGUUCGGAAUUAAAAUUAGAAACGCCAAUGCUCUUGGUGAGGCCCAGCUCCACC ....((.(((((((........)))))))((((((.(((..(((.(((((......(((((.(....))))))(((....)))......))))))))....))).)))))).))...... ( -33.40, z-score = -0.15, R) >dp4.chrXR_group8 587577 120 + 9212921 AACCGCAAGCGACUCACCUGAUUGUGGAUUGGUUUGAUUUUGCAAUUGGCCAGCAGGCGCGUCAGCUGCUCGGAAUUGAAAUUGGAAACGCCGAUACUUUUGGUGAGGCCCAGCUCGACC ..(((..((((((.(.((((...((.(((((...........))))).))...)))).).))).)))...)))..((((..(.((...((((((.....))))))...)).)..)))).. ( -34.90, z-score = 0.40, R) >droPer1.super_32 95067 120 - 992631 AACCGCAAUCGACUCACCUGAUUGUGGAUUGGCUUGAUUUUGCAAUUGGCCAGCAGGCGCGUCAGCUGCUCGGAAUUGAAAUUGGAAACGCCGAUACUUUUGGUGAGGCCCAGCUCGACC ..(((((((((.......)))))))))...(((((...(((.(((((..(((((((.(......)))))).))......))))).)))((((((.....))))))))))).......... ( -37.40, z-score = -0.49, R) >droWil1.scaffold_180698 2107908 94 + 11422946 --------------------------AACCGGCUUUAUUUUGGCCACGUCAAGCAAUCGAUUGAUUUGCUGUGCAUUGAAGUUGGAGACACCAAUCCCACGACAGAGGCCAGCAUCGACC --------------------------....((((.......))))..(((....((((....))))(((((.((.(((..((.(((........))).))..)))..)))))))..))). ( -24.30, z-score = -0.89, R) >droVir3.scaffold_12930 29285 120 - 74342 AACUCAGCUUGCCUUACCUGGUUGUGAAUUGGCUUGAUUUUGCAGUUGGCCAAGAGGCGCGUCAGUUGCUCGGAAUUGAAAUUGGAAACGCCAAUGCUCUUCGUCAAGCCGAGCUCGACA ...((((((((((......)))........((((((((...(((.(((((...........((((((......))))))....(....))))))))).....))))))))))))).)).. ( -35.50, z-score = 0.21, R) >droMoj3.scaffold_6680 22687141 120 - 24764193 AACCGCAGUUGCCUAACCUGGUUAUGAAUCGGCUUGAUUUUGCAGUUGGCUAAGAGGCGCGUCAGCUGCUCGGAAUUGAAAUUGGAAACGCCAAUGCUUUUGGUCAGGCCCAGCUCGACG ...((.(((((((......)).........(((((((((..(((.(((((...(((..((....))..)))............(....)))))))))....))))))))))))))))... ( -36.70, z-score = 0.12, R) >droGri2.scaffold_15110 480950 112 + 24565398 --------AAAUACAACCUGAUUGUGUAUUGGCUUGAUUUUGCAGUUGGCCAACAGACGCUUUAGCUGCUCGGAAUUGAAAUUGGAGACGCCGAUGUUUUUGACCAGGCCUAAAUCGAUC --------...........(((((......((((((.....(((((((((........)))..))))))((((((.....(((((.....)))))..)))))).)))))).....))))) ( -27.80, z-score = -0.82, R) >apiMel3.Group15 4736071 106 + 7856270 --------------UACUUGAUUCAUGACAGGUUUGAUUUGAGCGAUUGAUAAAAUACGAUCGAUCUGUUGGGAAUUAAAAUUACUAAGACCUAUGCUUUUCGUUAAUCCCAAUUUUACA --------------..((..(.(((.........))).)..)).(((((((........))))))).(((((((.((((.......(((.......)))....)))))))))))...... ( -18.40, z-score = -0.55, R) >triCas2.ChLG1X 524079 99 - 8109244 ---------------------UUGACAACGGGCUUGAUUUUACAAUUUUUGAUGAGACGCUCGAUUUGUUCCUCGUUGAAGUUGGACACUCCGAUGCUUUUUGUGAGACCCAGACGGACG ---------------------........(((.....((((((((..((..(((((..((.......))..)))))..))(((((.....))))).....)))))))))))......... ( -22.00, z-score = -0.23, R) >consensus AACCACAAUUGACGCACCUGAUUGUGGAUUGGCUUGAUUUUGCAGUUGGCCAGCAGGCGAGUCAGCUGCUCGGAAUUGAAAUUGGAAACGCCGAUGCUCUUGGUCAGGCCCAGUUCGACC ..............................(((((.(((..(((((((((..........)))))))))...........(((((.....)))))......))).))))).......... (-16.01 = -15.84 + -0.17)

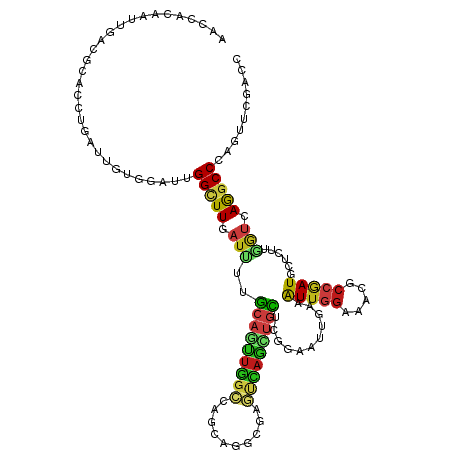

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:49 2011