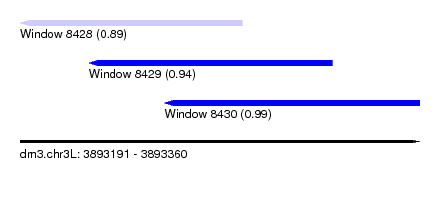
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,893,191 – 3,893,360 |
| Length | 169 |
| Max. P | 0.992366 |

| Location | 3,893,191 – 3,893,285 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.82 |
| Shannon entropy | 0.60447 |
| G+C content | 0.66317 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -18.00 |
| Energy contribution | -17.09 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3893191 94 - 24543557 UGCGGACGCAUUGCGCCG-----------CAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUUCUCCGGGGAUAGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGA--- (((((.(((...))))))-----------))((((((.......))))))..(((((((....))))))).....((((((((.(((......))))))))))).--- ( -51.00, z-score = -2.59, R) >droSim1.chr3L_random 193150 92 - 1049610 UGCGGACGCAUUGCGCCG-----------CAGGAGCGUCUCCUUCGCUCCGCCCCUGGACU--UCCGGGGGUGGGCGGGGGCGAGCGUUCUAUUGCCGCCUUCGA--- (((((.(((...))))))-----------))((((((((.(((((((.((((((((((...--.))))))))))))))))).)).))))))..............--- ( -55.60, z-score = -3.59, R) >droSec1.super_2 3886401 92 - 7591821 UGCGGACGCAUUGCGCCG-----------CAGGAGCGUCUCCUUCGCUCCGCCCCUGGACU--ACCGGGGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGA--- (((((.(((...))))))-----------))((((((((.(((((((.((((((((((...--.))))))))))))))))).)).))))))..............--- ( -57.20, z-score = -3.92, R) >droYak2.chr3L 4452267 92 - 24197627 UGCAGACGCAUUGCGCCG-----------CAGGAGCGUCUCCUUCGCUCCGCUCCUGGAUU--UCCAGUGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGA--- .((((.....))))((.(-----------((((((((((.(((((((.((((..((((...--.))))..))))))))))).)).))))))..))).))......--- ( -45.80, z-score = -2.11, R) >droEre2.scaffold_4784 6591914 92 - 25762168 UGCAGACGCAUUGCGCCG-----------CAGGAGCGUCUCCUUCGCUCCGCCCCCGGACC--UCCGGUGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGA--- .((((.....))))((.(-----------((((((((((.(((((((.(((((.((((...--.)))).)))))))))))).)).))))))..))).))......--- ( -48.50, z-score = -1.92, R) >droWil1.scaffold_180955 1850527 108 - 2875958 UGCAGACGCAUUGCACUGUUUGGCCCAUCCGUACGCCGCUUCUCCCCUCUCCUCACGGUUGGGGGCUGGAGUGGUGGGGGGCGUGCGUGAAAUUGCCGCCCUUUGGAU ..((((((........))))))....(((((..(((((((((.(((((..((....))..)))))..)))))))))(((((((.(((......))))))))))))))) ( -49.00, z-score = -1.52, R) >droVir3.scaffold_13049 21118795 75 + 25233164 UGCAGACGCAUUGCGCC------------GUUGGCUCAUCCAUACGCUCU---------------UCG---UGGGUGGGGGCGUGCGCUACAUUGCCGCCUUUGA--- ..((((.(((..(((((------------(((..(((((((((.......---------------..)---)))))))))))).)))).....)))....)))).--- ( -23.90, z-score = 1.21, R) >droMoj3.scaffold_6680 22237259 75 + 24764193 UGCGGACGCAUUGUGCC------------GUUUGCCCAUUCCUACGCUCU---------------UCG---UGGGCGGGGGCGCGCGUCGAAUUGCCGUCUUCGA--- .(.(((((((...((.(------------((.(((((...((((((....---------------.))---))))...))))).))).))...)).))))).)..--- ( -30.40, z-score = -0.57, R) >droGri2.scaffold_15110 10674467 78 - 24565398 UGCAGACGCAUUGCGUC------------AUUGGCCCAAUCCUACGUUCC---------------UUGAAUGGGGCGGGGGCGUGCGCUGAAUUGCCGCCUUUGU--- .((((((((...)))))------------.(..((.((..((..((((((---------------......))))))..))..)).))..)..))).........--- ( -27.90, z-score = -0.01, R) >consensus UGCAGACGCAUUGCGCCG___________CAGGAGCGUCUCCUUCGCUCCGCCCCUGGACU__UCCGGGGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGA___ .((((.....))))...............................((((.......................))))(((((((.(((......))))))))))..... (-18.00 = -17.09 + -0.91)
| Location | 3,893,220 – 3,893,323 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 66.05 |
| Shannon entropy | 0.65737 |
| G+C content | 0.61039 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -18.30 |
| Energy contribution | -16.08 |
| Covariance contribution | -2.23 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3893220 103 - 24543557 GUGCUCGUC-CUGGCACGUUUCCGAUCAC-GAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUUCUCCGGGGAUAGGC------ ..((.((((-(..(((((((((.......-)))))....)))))))))....))(((((.((((((.......))))))))(((((((....)))))))...)))------ ( -43.90, z-score = -2.10, R) >droSim1.chr3L_random 193179 101 - 1049610 GUGCUCGUC-CUGGCACGUUUCCGAUCAC-GAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACU--UCCGGGGGUGGGC------ (((((....-..)))))(((((.......-))))).....(((((.(((...))))))))((((((.......))))))((((((((...--.))))))))....------ ( -47.70, z-score = -2.43, R) >droSec1.super_2 3886430 101 - 7591821 GUGCUCGUC-CUGGCACGUUUCCGAUCAC-GAAACUGAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACU--ACCGGGGGUGGGC------ (((((....-..)))))(((((.......-))))).(((.(((((.(((...))))))))((((...)))))))((.((((((((((...--.))))))))))))------ ( -47.30, z-score = -2.16, R) >droYak2.chr3L 4452296 101 - 24197627 GCGCUCGUC-CUGGCACGUUCCCGAUCAC-GGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCUCCUGGAUU--UCCAGUGGUGGGC------ ((((.((((-...(((((((((.......-)))))....)))).))))....)))).(.(((((...))))).)((.((((..((((...--.))))..))))))------ ( -41.80, z-score = -1.50, R) >droEre2.scaffold_4784 6591943 101 - 25762168 GCGCUCGUC-CUGGCACGUUCCCGAUCAC-GGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCCGGACC--UCCGGUGGUGGGC------ ((((.((((-...(((((((((.......-)))))....)))).))))....)))).(.(((((...))))).)((.(((((.((((...--.)))).)))))))------ ( -44.50, z-score = -1.43, R) >droWil1.scaffold_180955 1850564 109 - 2875958 GUGCUCGUC-CUGGCACAUUUCCUAUCCU-GGAAUAUUAGUGCAGACGCAUUGCACUGUUUGGCCCAUCCGUACGCCGCUUCUCCCCUCUCCUCACGGUUGGGGGCUGGAG ((((.((((-...((((..((((......-)))).....)))).))))....)))).....(((..........))).((((.(((((..((....))..)))))..)))) ( -35.60, z-score = -1.07, R) >droVir3.scaffold_13049 21118824 84 + 25233164 GCGCUCGUC-UUAGCACGUUCCA-AUGUUAGGAGCUUGAUUGCAGACGCAUUGCGCCG-UUGGCUCAUCCAUACGCUCUUCG--UGGGU---------------------- ((((.((((-((((((.(((((.-......))))).)).))).)))))....)))).(-((((.....))).))((((....--.))))---------------------- ( -23.60, z-score = 0.43, R) >droMoj3.scaffold_6680 22237288 86 + 24764193 GUGCUCGUCUUUGGCGCUUUCCGUAUUUUACGAGAAACAGUGCGGACGCAUUGUGCCG-UUUGCCCAUUCCUACGCUCUUCG--UGGGC---------------------- ......((...((((...(((((((...)))).)))(((((((....)))))))))))-...)).....((((((.....))--)))).---------------------- ( -24.20, z-score = -0.18, R) >droGri2.scaffold_15110 10674496 87 - 24565398 GUGCUCGUC-CUGGCAUGUUCCC-GUAUUAGGAACUUAAAUGCAGACGCAUUGCGUCA-UUGGCCCAAUCCUACGUUCCUUGAAUGGGGC--------------------- (((((....-..)))))..((((-((...((((((.........(((((...))))).-..((......))...))))))...)))))).--------------------- ( -26.20, z-score = -1.11, R) >consensus GUGCUCGUC_CUGGCACGUUCCCGAUCAC_GGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACU__UCCGGGGGUGGGC______ ((((.((((....(((((((((........)))))....)))).))))....))))....(((.....))).....(((......)))....................... (-18.30 = -16.08 + -2.23)


| Location | 3,893,252 – 3,893,360 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 58.52 |
| Shannon entropy | 0.90300 |
| G+C content | 0.51937 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -18.12 |
| Energy contribution | -16.39 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3893252 108 - 24543557 ACUCCCAAUAUAUUUCAGGAGAA-GAGCUUUAAAAAGGGUGCUCGUC-CUGGCACGUUUCCGAUCAC-GAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUU--- ................((((((.-..(((((......(((((.((((-(..(((((((((.......-)))))....)))))))))....)))))...))))).)))))).--- ( -34.70, z-score = -1.46, R) >droSim1.chr3L_random 193209 108 - 1049610 ACUCCCACUAUAUUUCUCGGAAA-GAGCUUUAAAAAGGGUGCUCGUC-CUGGCACGUUUCCGAUCAC-GAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUU--- .((((.......(((((((((((-((((............))))((.-...))...)))))))....-))))......(((((.(((...)))))))))))).........--- ( -30.80, z-score = -0.34, R) >droSec1.super_2 3886460 108 - 7591821 ACUCCCACUAUAUUUCUCGGAAA-GAGCUUUAAAAAGGGUGCUCGUC-CUGGCACGUUUCCGAUCAC-GAAACUGAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUU--- .((((.(((.(((((((((((((-((((............))))((.-...))...)))))))....-)))).)).)))((((.(((...))))))).)))).........--- ( -31.00, z-score = -0.13, R) >droYak2.chr3L 4452326 107 - 24197627 ACCCCCACUAUAUGUGGAAUGGA-GAGCUUU-AAAAGGGCGCUCGUC-CUGGCACGUUCCCGAUCAC-GGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUU--- ....((((.....))))...(((-(((((((-.....(((((.((((-...(((((((((.......-)))))....)))).))))....)))))...))))).)))))..--- ( -41.60, z-score = -2.51, R) >droEre2.scaffold_4784 6591973 107 - 25762168 ACACCCACUACAGUGGCCACAUA-GAGUUUU-AAAAGGGCGCUCGUC-CUGGCACGUUCCCGAUCAC-GGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUU--- ....((((....)))).......-.......-.....(((((.((((-...(((((((((.......-)))))....)))).))))....)))))..(((((...))))).--- ( -33.40, z-score = -0.81, R) >droAna3.scaffold_13337 13748792 104 + 23293914 --ACUUUGUCCACAAACUUGGCA-AAUGUGUUAAAAGGGUGCUCGUC-UUGGCGCGUU--CGAUCGC-UGAACAAAAGUGCAGACGCAUUGCGCCGCAGGACCAUCUCAUU--- --((((((.(((......)))))-)).)).......(((((...(((-((((((((((--((.....-)))))...(((((....)))))))))))..)))))))))....--- ( -29.80, z-score = -0.17, R) >dp4.chrXR_group6 4962441 97 - 13314419 ----ACCUCCCGCUAAACUGCCUCUUGCUUCAAAAAGGGUAUCCGA--GUCAGGUGUUCGCAA-----CGAAAGCGCGCCGAGUCGCACUGCGCCUUUGGGACCAUCU------ ----...((((((......))............((((((((..(((--.((.((((..(((..-----.....))))))))).)))...))).)))))))))......------ ( -27.00, z-score = 0.17, R) >droPer1.super_9 3265166 96 - 3637205 ----ACCUCCCGCUAAAUUGCCUCUUGCAUCAAAAAGGGUAUCCGA--GUCAGGUGUUCGCAA-----CGAAAGCGCGCCGAGUCGCACUGCGCC-UUGGGACCAUCU------ ----...((((((......)).............(((((((..(((--.((.((((..(((..-----.....))))))))).)))...))).))-))))))......------ ( -26.10, z-score = 0.34, R) >droWil1.scaffold_180955 1850599 111 - 2875958 ACAUUUUCCACAUGAGCAUCGAA-AUCUUCAGAAAAGGGUGCUCGUC-CUGGCACAUUUCCUAUCCU-GGAAUAUUAGUGCAGACGCAUUGCACUGUUUGGCCCAUCCGUACGC .........(((((((((((...-.((....))....))))))))).-..(((.((.((((......-))))...((((((((.....))))))))..))))).....)).... ( -27.50, z-score = -0.58, R) >droVir3.scaffold_13049 21118838 104 + 25233164 AUAUGCGUCUCGUGUCA-GGUCU---CAAUUAAAAAGGGCGCUCGUC-UUAGCACGUUCCA-AUGUUAGGAGCUUGAUUGCAGACGCAUUGCGCCGUUGGCUCAUCCAUA---- ..((((((((...((((-((.((---(.........((((....)))-)(((((.......-.))))).)))))))))...))))))))((.(((...))).))......---- ( -29.90, z-score = -0.70, R) >droMoj3.scaffold_6680 22237302 110 + 24764193 AUAUGCAGCUCAUUUUCGGAGUUGGGUUAUUAAAAAGGGUGCUCGUCUUUGGCGCUUUCCGUAUUUUACGAGAAACAGUGCGGACGCAUUGUGCCGUUUGCCCAUUCCUA---- .................(((((.((((......(((.((..(.(((((...((((((((((((...)))).)))..))))))))))....)..)).))))))))))))..---- ( -33.10, z-score = -1.30, R) >droGri2.scaffold_15110 10674513 105 - 24565398 AUACGCACAUUGUUUUGUGCUCG---CAUUUAAAAAGGGUGCUCGUC-CUGGCAUGUUCCC-GUAUUAGGAACUUAAAUGCAGACGCAUUGCGUCAUUGGCCCAAUCCUA---- ....(((((......)))))..(---(((((((.....(((((....-..)))))(((((.-......))))))))))))).(((((...)))))...............---- ( -31.30, z-score = -1.79, R) >consensus ACACCCACUACAUUUCACGGGCA_GAGCUUUAAAAAGGGUGCUCGUC_CUGGCACGUUCCCGAUCAC_GGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCCCCUU___ .....................................(((((.((((....(((((((((........)))))....)))).))))....)))))................... (-18.12 = -16.39 + -1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:47 2011