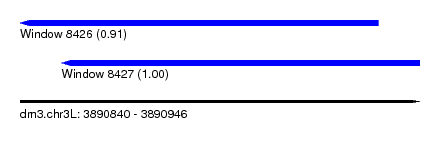
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,890,840 – 3,890,946 |
| Length | 106 |
| Max. P | 0.997279 |

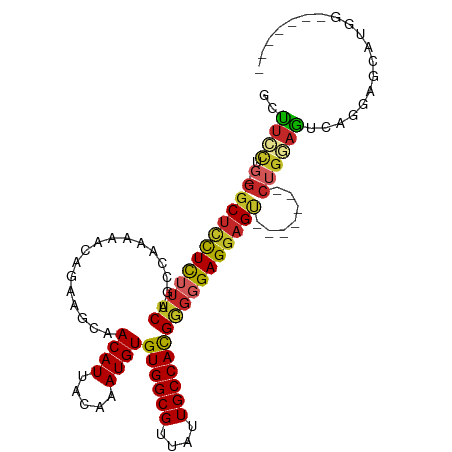
| Location | 3,890,840 – 3,890,935 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.90 |
| Shannon entropy | 0.50761 |
| G+C content | 0.47428 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -18.53 |
| Energy contribution | -19.34 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
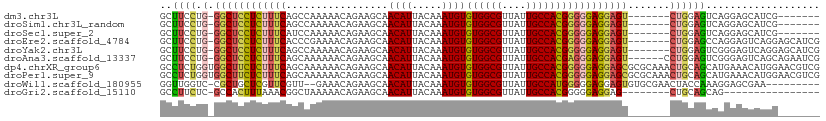
>dm3.chr3L 3890840 95 - 24543557 UCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCAGGAGC-------AUCGACUUCUUUUUG- ((((((..(.................((((.....))))((((((....)))))))..))))))..-------..((((((......-------...))))))......- ( -29.50, z-score = -1.86, R) >droSim1.chr3L_random 190814 95 - 1049610 UCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCAGGAGC-------AUCGACUUCUUUUUG- ((((((..(.................((((.....))))((((((....)))))))..))))))..-------..((((((......-------...))))))......- ( -29.50, z-score = -1.86, R) >droSec1.super_2 3884092 95 - 7591821 UCCUCUUUCAUCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCAGGAGC-------AUCGACUUCUUUUUG- ((((((..(.................((((.....))))((((((....)))))))..))))))..-------..((((((......-------...))))))......- ( -29.50, z-score = -2.11, R) >droEre2.scaffold_4784 6589579 102 - 25762168 UCCUCUUUCACCCGAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGCCAGGAGUCAGGAGCAUCGACUUCUUUUUG- ..(((((((.(((.............((((.....))))((((((....)))))))))))))))).-------........(((((((.........))))))).....- ( -31.20, z-score = -1.04, R) >droYak2.chr3L 4448882 102 - 24197627 UCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCGGGAGUCAGGAGCAUCGACUUCUUUUUG- ((((((..(.................((((.....))))((((((....)))))))..))))))..-------..((((((((..((.....)).))))))))......- ( -33.60, z-score = -2.04, R) >droAna3.scaffold_13337 13746113 104 + 23293914 UCCUCUUUCAGCAAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGAGGGAGGAGU------CCUGGAGUCGGGAGUCAGCAGAAUCGACUUAUUUUUUG (((((((((.................((((.....))))((((((....)))))))))))))))..------....(((((((...((....)).)))))))........ ( -32.60, z-score = -2.53, R) >dp4.chrXR_group6 4960186 108 - 13314419 UUCUCUUUCAGCAAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGCGCGCAAACUGCAGCAUGAAACAUGGAACGUCGACUGCUUUUG-- ((((((..(.................((((.....))))((((((....)))))))..)))))).....((((..((((((((...))))(.....)..)))).))))-- ( -26.00, z-score = 0.29, R) >droPer1.super_9 3262925 108 - 3637205 UUCUCUUUCAGCAAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGCGCGCAAACUGCAGCAUGAAACAUGGAACGUCGACUGCUUUUG-- ((((((..(.................((((.....))))((((((....)))))))..)))))).....((((..((((((((...))))(.....)..)))).))))-- ( -26.00, z-score = 0.29, R) >droWil1.scaffold_180955 1847886 98 - 2875958 UGCUCGUUCGUU--GAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCAUGGGGGAGGAGUGUGCGAACUACCA-AAGGAGC-----GAAACAUCUUUUUG---- ...((((((.((--.........(((.((((.....(.(((((((....))))))).).....))))))).........-)).))))-----))............---- ( -24.47, z-score = -0.16, R) >droMoj3.scaffold_6680 22235219 74 + 24764193 -----------CAGAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGCA---GUACUCCACUCAACUCCA---------------------- -----------...............((((.....))))((((((....))))))(((((((.....---...)))).))).......---------------------- ( -19.90, z-score = -1.41, R) >consensus UCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU_______CUGGAGUCAGGAGC_______AUCGACUUCUUUUUG_ (((((((((.................((((.....))))((((((....))))))))))))))).............................................. (-18.53 = -19.34 + 0.81)

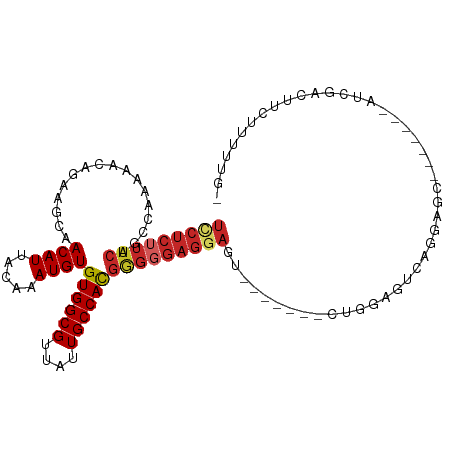
| Location | 3,890,851 – 3,890,946 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.15 |
| Shannon entropy | 0.46741 |
| G+C content | 0.52055 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.37 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3890851 95 - 24543557 GCUUCCUG-GGCUCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCAGGAGCAUCG------- ((((((..-(((((((((..(.................((((.....))))((((((....)))))))..))))))))-------)..).....)))))....------- ( -39.70, z-score = -3.82, R) >droSim1.chr3L_random 190825 95 - 1049610 GCUUCCUG-GGCUCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCAGGAGCAUCG------- ((((((..-(((((((((..(.................((((.....))))((((((....)))))))..))))))))-------)..).....)))))....------- ( -39.70, z-score = -3.82, R) >droSec1.super_2 3884103 95 - 7591821 GCUUCCUG-GGCUCCUCUUUCAUCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCAGGAGCAUCG------- ((((((..-(((((((((..(.................((((.....))))((((((....)))))))..))))))))-------)..).....)))))....------- ( -39.70, z-score = -4.15, R) >droEre2.scaffold_4784 6589590 102 - 25762168 GCUUCCUG-GGCUCCUCUUUCACCCGAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGCCAGGAGUCAGGAGCAUCG (((.((((-(.((((((((((.(((.............((((.....))))((((((....)))))))))))))))).-------(((....)))))))))))))).... ( -41.70, z-score = -3.00, R) >droYak2.chr3L 4448893 102 - 24197627 GCUUCCUG-GGCUCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU-------CUGGAGUCGGGAGUCAGGAGCAUCG ((((((..-(((((((((..(.................((((.....))))((((((....)))))))..))))))))-------)..).(.(....).).))))).... ( -42.20, z-score = -3.50, R) >droAna3.scaffold_13337 13746125 103 + 23293914 GCUUCCUG-GGCUCCUCUUUCAGCAAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGAGGGAGGAGU------CCUGGAGUCGGGAGUCAGCAGAAUCG ((((((.(-((((((((((((.................((((.....))))((((((....)))))))))))))))))------)).)))(.(....).))))....... ( -41.80, z-score = -3.88, R) >dp4.chrXR_group6 4960196 110 - 13314419 GCCUCUGGUGGCUUCUCUUUCAGCAAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGCGCGCAAACUGCAGCAUGAAACAUGGAACGUCG ((.....(((((((((((..(.................((((.....))))((((((....)))))))..)))))))).))).....))..((((...))))........ ( -34.20, z-score = -1.06, R) >droPer1.super_9 3262935 110 - 3637205 GCCUCUGGUGGCUUCUCUUUCAGCAAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGCGCGCAAACUGCAGCAUGAAACAUGGAACGUCG ((.....(((((((((((..(.................((((.....))))((((((....)))))))..)))))))).))).....))..((((...))))........ ( -34.20, z-score = -1.06, R) >droWil1.scaffold_180955 1847897 98 - 2875958 GGUUGGUC-CGCUGCUCGUUCGUU--GAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCAUGGGGGAGGAGUGUGCGAACUACCAAAGGAGCGAA--------- ((((.(..-((((.(((.((((((--(.........))))...........((((((....))))))))).))).))))..).))))..............--------- ( -29.50, z-score = -0.55, R) >droGri2.scaffold_15110 10672307 85 - 24565398 GCCUUCUC-GCCACUUUAAACGGCUAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAG--------CUGCAGCAG---------------- ((((((((-.((..........(((.........))).((((.....))))((((((....)))))).)).))))))--------..)).....---------------- ( -24.90, z-score = -0.99, R) >consensus GCUUCCUG_GGCUCCUCUUUCAGCCAAAAACAGAAGCAACAUUACAAAUGUGUGGCGUUAUUGCCACGGGGGAGGAGU_______CUGGAGUCAGGAGCAUGG_______ ..........(((((((((((.................((((.....))))((((((....)))))))))))))))))................................ (-24.30 = -24.37 + 0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:44 2011