| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,889,794 – 3,889,897 |
| Length | 103 |
| Max. P | 0.535245 |

| Location | 3,889,794 – 3,889,897 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 67.50 |
| Shannon entropy | 0.67163 |
| G+C content | 0.47310 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -8.70 |
| Energy contribution | -8.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3889794 103 - 24543557 GGCAAUCGCAAACAAAACGAGUGAGAAAAGCCUCACUCACU-CGAAUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUACGGCC-CACCAACCUCGA (((..............((((((((..........))))))-))......((((((((....(((((...)))))..))))))))....)))-............ ( -25.40, z-score = -1.55, R) >droSim1.chr3L_random 189809 103 - 1049610 GGCAAUCGCAAACAAAACGAGUGAGAAAAGCCUCACUCCCU-CGAAUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUACGGCC-CACCAACCUCGC (((...............(((((((......)))))))...-........((((((((....(((((...)))))..))))))))....)))-............ ( -24.00, z-score = -1.22, R) >droSec1.super_2 3883064 103 - 7591821 GGCAAUCGCAAACAAAACGAGUGAGAAAAGCCUCACUCCCU-CGAAUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUACGGCC-CACCAACCUCGC (((...............(((((((......)))))))...-........((((((((....(((((...)))))..))))))))....)))-............ ( -24.00, z-score = -1.22, R) >droEre2.scaffold_4784 6588513 78 - 25762168 -------------------------GGCAAUCGCAAACACU-CGAAUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUACGGCC-CACCAGCCUCGA -------------------------(((........(((((-(.........))))))(((.(((((...))))).))))))......(((.-.....))).... ( -17.90, z-score = -0.83, R) >droYak2.chr3L 4447847 78 - 24197627 -------------------------GGCAAUCGCAAACACU-CGAGUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUACGGCC-CACCAGCCUCGA -------------------------(((........(((((-(.........))))))(((.(((((...))))).))))))......(((.-.....))).... ( -17.90, z-score = -0.45, R) >droAna3.scaffold_13337 13745069 95 + 23293914 --CAACAGCAAACAAUUGGUGUGAGGAACCCCUCGC--ACU-CAAGUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUAUGGCC-CACCAACC---- --.............(((((((((((....))))))--)).-)))........((((.(((.(((((...))))).)))(((......))).-))))....---- ( -27.40, z-score = -1.92, R) >dp4.chrXR_group6 4959111 78 - 13314419 -------------------------GACACGCACACACACU-CAAGUCUCAAGGGGGUGUUCAAUUGUGUCAAUUUAACGCCUUUUAU-GCCACACCAACUCUAA -------------------------((((((.....(((((-(...........)))))).....)))))).................-................ ( -13.00, z-score = -0.12, R) >droPer1.super_9 3261849 78 - 3637205 -------------------------GACACGCACACACACU-CAAGUCUCAAGGGGGUGUUCAAUUGUGUCAAUUUAACGCCUUUUAU-GCCACACCAACUCUAA -------------------------((((((.....(((((-(...........)))))).....)))))).................-................ ( -13.00, z-score = -0.12, R) >droMoj3.scaffold_6680 22233945 93 + 24764193 -----GGGAGAGGGAGAGGCAGAGAGAAUGCCUCUCGCACA-CAAAUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUAUGGCAACCCCAA------ -----..(((((.((((((((.......)))))))).)...-....))))..(((.(((((.(((((...))))).)))(((......))).)))))..------ ( -32.01, z-score = -2.35, R) >droGri2.scaffold_15110 10671000 104 - 24565398 AACAACAGGCAACGUUUGGGAAACUUCGAACGGCGUUUACAGUAAAUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUAGGACA-CCCCGCCAACGA .......(((..(((((((....)..))))))....................(((.((((((((..((((.......))))...)).)))))-)))))))..... ( -27.60, z-score = -0.87, R) >consensus _______G__AAC_A___G______GAAAGCCUCACACACU_CAAAUCUCAAGGGUGUGUUCAAUUGUGUCAAUUUAACGCCUUUUACGGCC_CACCAACCUCGA ..................................................(((((.(((((.(((((...))))).))))))))))................... ( -8.70 = -8.70 + 0.00)


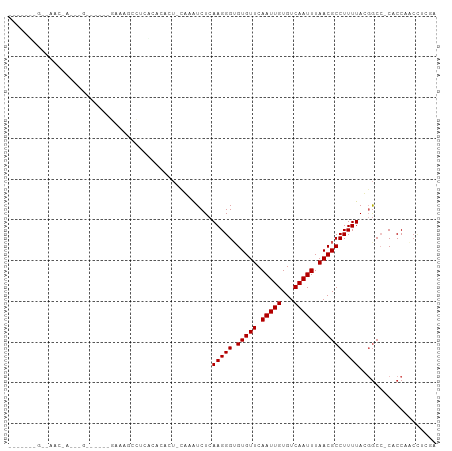
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:43 2011