
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,881,720 – 3,881,816 |
| Length | 96 |
| Max. P | 0.715415 |

| Location | 3,881,720 – 3,881,816 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 70.49 |
| Shannon entropy | 0.60404 |
| G+C content | 0.40073 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -7.47 |
| Energy contribution | -7.89 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715415 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
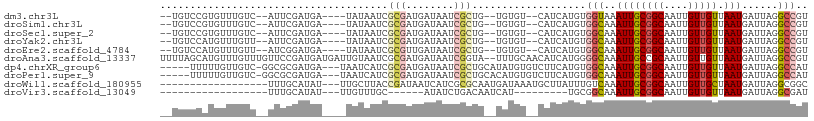
>dm3.chr3L 3881720 96 - 24543557 --UGUCCGUGUUUGUC--AUUCGAUGA----UAUAAUCGCGAUGAUAAUCGCUG--UGUGU--CAUCAUGUGGUAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCGU --.......((((..(--((..(((((----((((...(((((....)))))..--)))))--))))..)))..)))).(((((....(((......)))...))))) ( -30.90, z-score = -2.64, R) >droSim1.chr3L 3422557 96 - 22553184 --UGUCCGUGUUUGUC--AUUCGAUGA----UAUAAUCGCGAUGAUAAUCGCUG--UGUGU--CAUCAUGUGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCGU --.......(((((((--((..(((((----((((...(((((....)))))..--)))))--))))..))))))))).(((((....(((......)))...))))) ( -34.80, z-score = -3.48, R) >droSec1.super_2 3875007 96 - 7591821 --UGUCCGUGUUUGUC--AUUCGAUGA----UAUAAUCGCGAUGAUAAUCGCUG--UGUGU--CAUCAUGUGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCGU --.......(((((((--((..(((((----((((...(((((....)))))..--)))))--))))..))))))))).(((((....(((......)))...))))) ( -34.80, z-score = -3.48, R) >droYak2.chr3L 4440108 96 - 24197627 --UGUCCAUGUUUGUU--AUUCGAUGA----UAUAAUCGCGAUGAUAAUCGCUG--UGUGU--CAUCAUGUGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCGU --.......(((((((--((..(((((----((((...(((((....)))))..--)))))--))))..))))))))).(((((....(((......)))...))))) ( -32.70, z-score = -3.05, R) >droEre2.scaffold_4784 6580851 96 - 25762168 --UGUCCAUGUUUGUU--AUCGGAUGA----UAUAAUCGCGUUGAUAAUCGCUG--UGUGU--CAUCAUGUGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCGU --.......(((((((--((..(((((----((((...(((........)))..--)))))--))))..))))))))).(((((....(((......)))...))))) ( -29.10, z-score = -1.80, R) >droAna3.scaffold_13337 13736870 106 + 23293914 UUUUAGCAUGUUUGUUUGUUCCGAUGAUGAUUGUAAUCGCGAUGAUAAUCGGUA--UUUGCAACAUCAUGGGGCAAAUUGCCGCAAUUGUUGUUAAUGAUUAGGCCGU .............(((((((((.((((((.((((((.(.((((....)))))..--.))))))))))))))))))))).(((..(((..(.....)..))).)))... ( -30.30, z-score = -1.38, R) >dp4.chrXR_group6 4949067 99 - 13314419 -----UUUUUGUUGUC-GGCGCGAUGA---UAAUCAUCGCGAUGAUAAUCGCUGCAUAUGUGUCUUCAUGUGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCAU -----...........-(((((((((.---....))))))((..(((((.((((((....((((.......))))...)))))).)))))..)).........))).. ( -35.60, z-score = -2.88, R) >droPer1.super_9 3251859 99 - 3637205 -----UUUUUGUUGUC-GGCGCGAUGA---UAAUCAUCGCGAUGAUAAUCGCUGCACAUGUGUCUUCAUGUGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCAU -----...........-(((((((((.---....))))))((..(((((.((((((....((((.......))))...)))))).)))))..)).........))).. ( -34.90, z-score = -2.48, R) >droWil1.scaffold_180955 1832526 87 - 2875958 ------------------UUUGCAUAU---UUGCUUACCGAUAAUCAUCGCGCAAUGAUAAAUGCUUAUUUGUCAAAUUGCGGCAAUUGUUGCUAAUGAUUAGGCGGC ------------------...(((...---.)))...(((.(((((((..(((((((((((((....))))))))..)))))(((.....)))..)))))))..))). ( -24.10, z-score = -1.52, R) >droVir3.scaffold_13049 21105610 72 + 25233164 ------------------UUUGCAUAU---UUGUUUGC------AUAUCUGACAAUCAU---------UGCGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCGAU ------------------.........---....((((------..((((((((((.((---------(((.((.....)).))))).)))))))..)))...)))). ( -21.10, z-score = -1.88, R) >consensus __UGUCCAUGUUUGUC__AUUCGAUGA____UAUAAUCGCGAUGAUAAUCGCUG__UGUGU__CAUCAUGUGGCAAAUUGCGGCAAUUGUUGUUAAUGAUUAGGCCGU ......................................(((........)))...................(((..((((((((....))))).)))......))).. ( -7.47 = -7.89 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:40 2011