
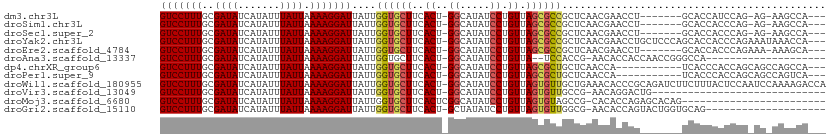
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,880,572 – 3,880,670 |
| Length | 98 |
| Max. P | 0.615613 |

| Location | 3,880,572 – 3,880,670 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.82 |
| Shannon entropy | 0.53265 |
| G+C content | 0.43491 |
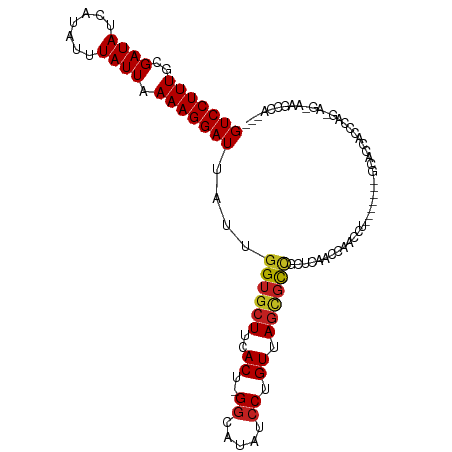
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -11.57 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3880572 98 - 24543557 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGCGCCGCUCAACGAACCU-------GCACCAUCCAG-AG-AAGCCA--- (((((((..((((.......)))).)))))))...((((((....((-((((.....))))))...((.....)).....-------)))))).....-..-......--- ( -20.50, z-score = -0.23, R) >droSim1.chr3L 3421394 98 - 22553184 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGCGCCGCUCAACGAACCU-------GCACCACCCAG-AG-AAGCCA--- (((((((..((((.......)))).))))))).....(.(((((.((-((........((((((...))).)))(.....-------.).....))))-.)-))))).--- ( -20.60, z-score = -0.38, R) >droSec1.super_2 3873872 98 - 7591821 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGCGCCGCUCAACGAACCU-------GCACCACCCAG-AG-AAGCCA--- (((((((..((((.......)))).))))))).....(.(((((.((-((........((((((...))).)))(.....-------.).....))))-.)-))))).--- ( -20.60, z-score = -0.38, R) >droYak2.chr3L 4438916 107 - 24197627 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGCGCCGCUCAACGAACCUGCUCCCAGCACCACCCAGAAAUAAACCA--- (((((((..((((.......)))).)))))))...(((((((.....-((((.....(((((((...))).))))....))))...)))))))...............--- ( -23.20, z-score = -1.58, R) >droEre2.scaffold_4784 6579664 99 - 25762168 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGCGCCGCUCAACGAACCU-------GCACCACCCAGAAA-AAAGCA--- (((((((..((((.......)))).)))))))...((((((....((-((((.....))))))...((.....)).....-------))))))........-......--- ( -20.40, z-score = -0.57, R) >droAna3.scaffold_13337 13735683 87 + 23293914 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUA--UCCACCG-AACACCACCAACCGGGCCA-------------------- (((((((..((((.......)))).)))))))...(((((.(((..(-((.(((......))--))))..)-)))))))((....))....-------------------- ( -20.00, z-score = -1.34, R) >dp4.chrXR_group6 4947780 96 - 13314419 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGCGCUGCUCAACCA-----------UCACCCACCAGCAGCCAGCCA--- (((((((..((((.......)))).)))))))...(((((((..((.-((.....)).)).)))((((((......-----------.........))))))..))))--- ( -22.26, z-score = -1.01, R) >droPer1.super_9 3250574 96 - 3637205 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGCGCUGCUCAACCA-----------UCACCCACCAGCAGCCAGUCA--- (((((((..((((.......)))).))))))).(((((((((.....-)))))...........((((((......-----------.........))))))))))..--- ( -20.46, z-score = -0.64, R) >droWil1.scaffold_180955 1830558 110 - 2875958 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGUGUUGCUGAAACACCCGCAGAUCUUCUUUACUCCAAUCCAAAAGACCA (((((((..((((.......)))).))))))).(((((((((.....-))))....((((..(((((.....)))))..))))............)))))........... ( -23.20, z-score = -1.47, R) >droVir3.scaffold_13049 21104127 80 + 25233164 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GGCAUAUCCUGUUAGUGUUGCCG-AACAGGACUG----------------------------- (((((((..((((.......)))).)))))))..((((((...((((-((((.....)))))))).)))))-).........----------------------------- ( -20.70, z-score = -1.87, R) >droMoj3.scaffold_6680 22222532 86 + 24764193 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACUCGGCAUAUCCUGUUAGUGUAGCCG-CACACCAGAGCACAG------------------------ (((((((..((((.......)))).))))))).....((((((....((((((((.......)))).))))-.......))))))..------------------------ ( -20.90, z-score = -1.53, R) >droGri2.scaffold_15110 10662359 89 - 24565398 GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU-GCUAUAUCCUGUUAGUGUUGGCG-AACACCAGUACUGGUGCAG-------------------- (((((((..((((.......)))).)))))))((((((((.(((.((-((((........))))...)).)-)))))))))).........-------------------- ( -22.00, z-score = -1.13, R) >consensus GUCCUUUGCGAUAUCAUAUUUAUUAAAAGGAUUAUUGGUGCUUCACU_GGCAUAUCCUGUUAGCGCCGCUCAACCAACCU_______GCACCACCCAG_AG_AAGCCA___ (((((((..((((.......)))).)))))))....((((((..((............)).))))))............................................ (-11.57 = -11.28 + -0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:39 2011