
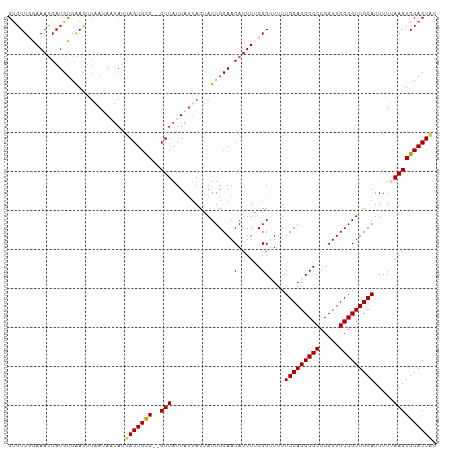
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,818,979 – 3,819,087 |
| Length | 108 |
| Max. P | 0.870178 |

| Location | 3,818,979 – 3,819,087 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 88.05 |
| Shannon entropy | 0.20508 |
| G+C content | 0.38744 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -23.38 |
| Energy contribution | -26.06 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3818979 108 + 24543557 GUGUUGGAAAUGAUGUGAACAUAAUAAUAUUAGUGCG--CUUAUUAUUACUAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUAAAGUGCACUAC ((((((((((((.((..(...((((((((..((....--))))))))))...)..)).)))))))).....(((((((((.....))))))))).........))))... ( -32.50, z-score = -2.48, R) >droEre2.scaffold_4784 6516802 108 + 25762168 GUGUUGAAAACGAUGUGGAUUUGAAAUUAUUAGUGCG--CUUAUUAUAACGAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUGCACUGC (..(((....)))..)..............(((((((--((((......((((((((((.....(((((.....)))))...))))))))))......))))))))))). ( -32.70, z-score = -1.94, R) >droYak2.chr3L 4375953 110 + 24197627 GCGUUGAAAAUGAUGUGGAUUGUAAAUGAAUAGUGUGUGCUUAUUACUAUAAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUAAAGUGCACUAC ((((((((((((.((..(.(((((....((((((....)).))))..))))))..)).)))))))))....(((((((((.....)))))))))........)))..... ( -31.00, z-score = -1.44, R) >droSec1.super_2 3812984 108 + 7591821 GUGUUCGAAAUGAUGUGAACUUAAUAAUAUUAGUGCG--CUUAUUAUUACUAUUGCAACAUUUACGGAUCUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUGCACUAC ..(((((........)))))..........(((((((--((((...............(......).....(((((((((.....)))))))))....))))))))))). ( -31.20, z-score = -2.44, R) >droSim1.chr3L 3359209 108 + 22553184 GUGUUCGAAAUGAUGUGAACUUAAUAAUAUAAGUGUG--CUUAUUAUUACUAUUGCAACAUUUACGGAUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUGCACUAC ((((((((((((.((..(...((((((((..((....--))))))))))...)..)).))))).)))....(((((((((.....))))))))).........))))... ( -30.80, z-score = -2.45, R) >consensus GUGUUGGAAAUGAUGUGAACUUAAUAAUAUUAGUGCG__CUUAUUAUUACUAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUGCACUAC ((((((((((((.((..(...((((((((............))))))))...)..)).)))))))).....(((((((((.....))))))))).........))))... (-23.38 = -26.06 + 2.68)


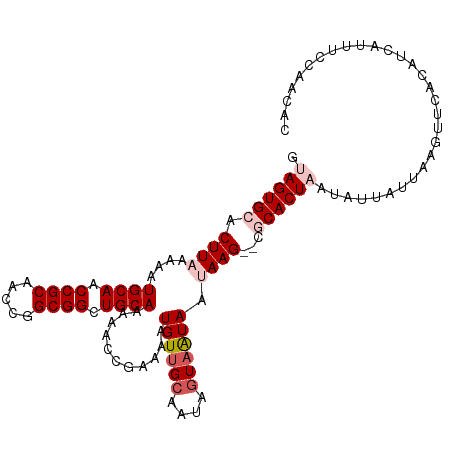
| Location | 3,818,979 – 3,819,087 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.05 |
| Shannon entropy | 0.20508 |
| G+C content | 0.38744 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -18.16 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3818979 108 - 24543557 GUAGUGCACUUUAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUAGUAAUAAUAAG--CGCACUAAUAUUAUUAUGUUCACAUCAUUUCCAACAC .((((((.(((.....((((.((((.....)))).))))...........((((((....))))))..)))--.)))))).............................. ( -25.50, z-score = -2.38, R) >droEre2.scaffold_4784 6516802 108 - 25762168 GCAGUGCACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUCGUUAUAAUAAG--CGCACUAAUAAUUUCAAAUCCACAUCGUUUUCAACAC ..(((((.((((....((((.((((.....)))).)))).........((((.......))))....))))--.)))))............................... ( -22.80, z-score = -1.53, R) >droYak2.chr3L 4375953 110 - 24197627 GUAGUGCACUUUAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUUAUAGUAAUAAGCACACACUAUUCAUUUACAAUCCACAUCAUUUUCAACGC ((.((((.........((((.((((.....)))).))))...........((((((.......)))))).)))))).................................. ( -22.20, z-score = -1.77, R) >droSec1.super_2 3812984 108 - 7591821 GUAGUGCACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAGAUCCGUAAAUGUUGCAAUAGUAAUAAUAAG--CGCACUAAUAUUAUUAAGUUCACAUCAUUUCGAACAC .((((((.((((....((((.((((.....)))).))))...........((((((....)))))).))))--.))))))..........((((..........)))).. ( -30.00, z-score = -3.61, R) >droSim1.chr3L 3359209 108 - 22553184 GUAGUGCACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAAAUCCGUAAAUGUUGCAAUAGUAAUAAUAAG--CACACUUAUAUUAUUAAGUUCACAUCAUUUCGAACAC ...(((.((((((...((((.((((.....)))).))))...........((((((....))))))(((((--....)))))....)))))).))).............. ( -24.80, z-score = -2.52, R) >consensus GUAGUGCACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUAGUAAUAAUAAG__CGCACUAAUAUUAUUAAGUUCACAUCAUUUCCAACAC .((((((.........((((.((((.....)))).))))...........((((((....))))))........)))))).............................. (-18.16 = -19.20 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:35 2011