
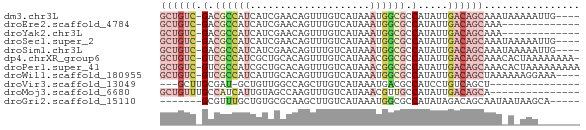
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,798,000 – 3,798,065 |
| Length | 65 |
| Max. P | 0.949554 |

| Location | 3,798,000 – 3,798,065 |
|---|---|
| Length | 65 |
| Sequences | 11 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 70.18 |
| Shannon entropy | 0.61064 |
| G+C content | 0.44605 |
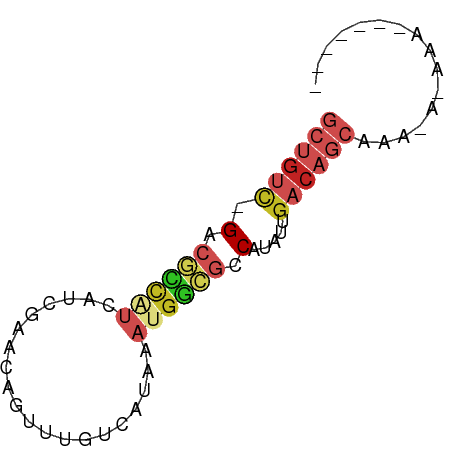
| Mean single sequence MFE | -16.56 |
| Consensus MFE | -6.76 |
| Energy contribution | -8.02 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949554 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
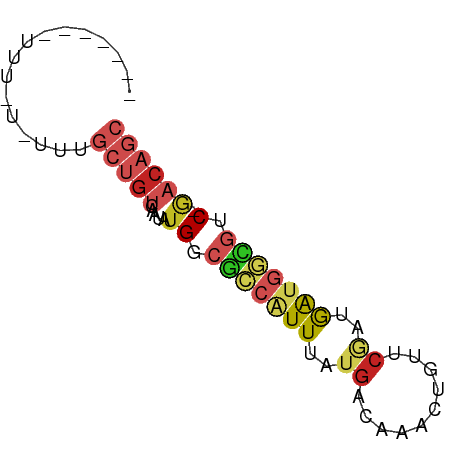
>dm3.chr3L 3798000 65 + 24543557 GCUGUC-GACGCCAUCAUCGAACAGUUUGUCAUAAAUGGCGCCAUAUUGACAGCAAAUAAAAAUUG---- ((((((-((((((((....((........))....)))))).....))))))))............---- ( -18.50, z-score = -3.12, R) >droEre2.scaffold_4784 6495527 56 + 25762168 GCUGUC-GACGCCAUCAUCGAACAGUUUGUCAUAAAUGGCGCCAUAUUGACAGCAAA------------- ((((((-((((((((....((........))....)))))).....))))))))...------------- ( -18.50, z-score = -3.13, R) >droYak2.chr3L 4354879 56 + 24197627 GCUGUC-GACGCCAUCAUCGAACAGUUUGUCAUAAAUGGCGCCAUAUUGACAGCAAA------------- ((((((-((((((((....((........))....)))))).....))))))))...------------- ( -18.50, z-score = -3.13, R) >droSec1.super_2 3792156 65 + 7591821 GCUGUC-GACGCCAUCAUCGAACAGUUUGUCAUAAAUGGCGCCAUAUUGACAGCAAAUAAAAAUUG---- ((((((-((((((((....((........))....)))))).....))))))))............---- ( -18.50, z-score = -3.12, R) >droSim1.chr3L 3338345 65 + 22553184 GCUGUC-GACGCCAUCAUCGAACAGUUUGUCAUAAAUGGCGCCAUAUUGACAGCAAAUAAAAAUUG---- ((((((-((((((((....((........))....)))))).....))))))))............---- ( -18.50, z-score = -3.12, R) >dp4.chrXR_group6 6857940 68 + 13314419 GCUGUC-GUCGCCAUCGCUGCACAGUUUGUCAUAAACGGCGCCAUAUUGACAGCAAACACUAAAAAAAA- ((((((-(.((((...(((....)))..((.....))))))......)))))))...............- ( -15.70, z-score = -1.20, R) >droPer1.super_41 216688 69 + 728894 GCUGUC-GUCGCCAUCGCUGCACAGUUUGUCAUAAACGGCGCCAUAUUGACAGCAAACACUAAAAAAAAA ((((((-(.((((...(((....)))..((.....))))))......)))))))................ ( -15.70, z-score = -1.19, R) >droWil1.scaffold_180955 1707224 65 + 2875958 GCUGUC-GUCGCCAUCAUUGCACAGUUUGUCAUAAAUGGCGCCAUAUUGACAGCUAAAAAAGGAAA---- ((((((-(.((((((...((.((.....))))...))))))......)))))))............---- ( -18.80, z-score = -2.42, R) >droVir3.scaffold_13049 21008698 51 - 25233164 ---GCUUGCGAU-GCUGUUGGCCAGCUUGUCAUAAAUGACGCCAUCCUGUCAGCU--------------- ---(((.(((..-((((.....))))..((((....)))).......))).))).--------------- ( -12.00, z-score = 0.16, R) >droMoj3.scaffold_6680 2641888 55 + 24764193 GCUGUUUGCCAUCAUUGUAGCCAAGUUUGUCAUAAACGUUGCCAUAUUGACAGCA--------------- ((((.......(((.(((.((...(((((...)))))...)).))).))))))).--------------- ( -10.11, z-score = -0.09, R) >droGri2.scaffold_15110 10574724 58 + 24565398 -------GCGUUUGCUGUGCGCAAGCUUGUCAUAAAUGGCGCCAUAUAGACAGCAAUAAUAAGCA----- -------((..(((((((((....))..((((....)))).........)))))))......)).----- ( -17.40, z-score = -1.02, R) >consensus GCUGUC_GACGCCAUCAUCGAACAGUUUGUCAUAAAUGGCGCCAUAUUGACAGCAAA_A_AAA_______ ((((((.(.((((((....................)))))).).....))))))................ ( -6.76 = -8.02 + 1.27)
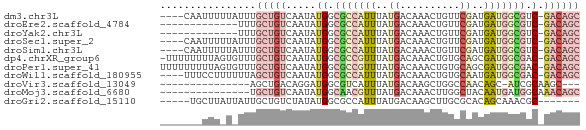
| Location | 3,798,000 – 3,798,065 |
|---|---|
| Length | 65 |
| Sequences | 11 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 70.18 |
| Shannon entropy | 0.61064 |
| G+C content | 0.44605 |
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -6.20 |
| Energy contribution | -6.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911356 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 3798000 65 - 24543557 ----CAAUUUUUAUUUGCUGUCAAUAUGGCGCCAUUUAUGACAAACUGUUCGAUGAUGGCGUC-GACAGC ----............((((((.....(((((((((...(((.....)))....)))))))))-)))))) ( -22.10, z-score = -3.66, R) >droEre2.scaffold_4784 6495527 56 - 25762168 -------------UUUGCUGUCAAUAUGGCGCCAUUUAUGACAAACUGUUCGAUGAUGGCGUC-GACAGC -------------...((((((.....(((((((((...(((.....)))....)))))))))-)))))) ( -22.10, z-score = -3.92, R) >droYak2.chr3L 4354879 56 - 24197627 -------------UUUGCUGUCAAUAUGGCGCCAUUUAUGACAAACUGUUCGAUGAUGGCGUC-GACAGC -------------...((((((.....(((((((((...(((.....)))....)))))))))-)))))) ( -22.10, z-score = -3.92, R) >droSec1.super_2 3792156 65 - 7591821 ----CAAUUUUUAUUUGCUGUCAAUAUGGCGCCAUUUAUGACAAACUGUUCGAUGAUGGCGUC-GACAGC ----............((((((.....(((((((((...(((.....)))....)))))))))-)))))) ( -22.10, z-score = -3.66, R) >droSim1.chr3L 3338345 65 - 22553184 ----CAAUUUUUAUUUGCUGUCAAUAUGGCGCCAUUUAUGACAAACUGUUCGAUGAUGGCGUC-GACAGC ----............((((((.....(((((((((...(((.....)))....)))))))))-)))))) ( -22.10, z-score = -3.66, R) >dp4.chrXR_group6 6857940 68 - 13314419 -UUUUUUUUAGUGUUUGCUGUCAAUAUGGCGCCGUUUAUGACAAACUGUGCAGCGAUGGCGAC-GACAGC -...............((((((.......(((((((..((((.....)).))..)))))))..-)))))) ( -18.20, z-score = -0.62, R) >droPer1.super_41 216688 69 - 728894 UUUUUUUUUAGUGUUUGCUGUCAAUAUGGCGCCGUUUAUGACAAACUGUGCAGCGAUGGCGAC-GACAGC ................((((((.......(((((((..((((.....)).))..)))))))..-)))))) ( -18.20, z-score = -0.61, R) >droWil1.scaffold_180955 1707224 65 - 2875958 ----UUUCCUUUUUUAGCUGUCAAUAUGGCGCCAUUUAUGACAAACUGUGCAAUGAUGGCGAC-GACAGC ----............((((((.......(((((((..((((.....)).))..)))))))..-)))))) ( -18.40, z-score = -2.36, R) >droVir3.scaffold_13049 21008698 51 + 25233164 ---------------AGCUGACAGGAUGGCGUCAUUUAUGACAAGCUGGCCAACAGC-AUCGCAAGC--- ---------------.((((...((..(((((((....))))..)))..))..))))-..(....).--- ( -13.70, z-score = -0.66, R) >droMoj3.scaffold_6680 2641888 55 - 24764193 ---------------UGCUGUCAAUAUGGCAACGUUUAUGACAAACUUGGCUACAAUGAUGGCAAACAGC ---------------((((((((.(.((((...((((.....))))...)))).).))))))))...... ( -14.40, z-score = -2.10, R) >droGri2.scaffold_15110 10574724 58 - 24565398 -----UGCUUAUUAUUGCUGUCUAUAUGGCGCCAUUUAUGACAAGCUUGCGCACAGCAAACGC------- -----.((........((((((((.(((....))).)).))).)))((((.....))))..))------- ( -11.90, z-score = 0.59, R) >consensus _______UUU_U_UUUGCUGUCAAUAUGGCGCCAUUUAUGACAAACUGUUCGAUGAUGGCGUC_GACAGC ................(((((....)))))((((((..((..........))..)))))).......... ( -6.20 = -6.20 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:30 2011