| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,787,983 – 3,788,105 |
| Length | 122 |
| Max. P | 0.977830 |

| Location | 3,787,983 – 3,788,090 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.45822 |
| G+C content | 0.56135 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -26.15 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.907881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
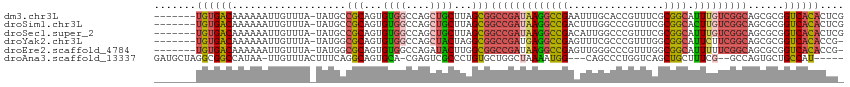
>dm3.chr3L 3787983 107 + 24543557 -------UGUGACAAAAAAUUGUUUA-UAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGAAUUUGCACCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCG -------...(((((....)))))..-.....((.(((((((((.(((((....(((((.....)))))................((((....)))))))))..))))))))))) ( -41.70, z-score = -1.81, R) >droSim1.chr3L 3328035 107 + 22553184 -------UGUGACAAAAAAUUGUUUA-UAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCACUUGUCGGCAGCGCGGUCACACUCG -------...(((((....)))))..-.....((.(((((((((.((((((....((((.....))))(((....(((((...)).)))....)))))))))..))))))))))) ( -45.70, z-score = -2.57, R) >droSec1.super_2 3782038 107 + 7591821 -------UGUGACAAAAAAUUGUUUA-UAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACAUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCG -------...(((((....)))))..-.....((.(((((((((.((((((....((((.....))))((((...(((((...)).)))...))))))))))..))))))))))) ( -47.40, z-score = -3.22, R) >droYak2.chr3L 4343910 106 + 24197627 -------UGUGACAAAAAAUUGUUUA-UAUGGCGCAGUGUGGCCAGCUACUAGGCGGCCGAUGAGGCCGAGUUUCGCCCGUUUGGCGGCAUUCUUCGGCAGCGCGGUCACACCG- -------((((((........(((..-...)))...((((.(((.(((....)))((((.....))))((((.(((((.....))))).))))...))).)))).))))))...- ( -39.80, z-score = -0.80, R) >droEre2.scaffold_4784 6483580 106 + 25762168 -------UGUGACAAAAAAUUGUUUA-UAUGGCGCAGUGUGGCCAGAUACUUGGCGGCCGAUAAGGCCGAGUUGGGCCCGUUUGGCGGCAUUUUUCGGCAGCGCGGUCACACCG- -------((((((........(((..-...)))...((((.(((.((......((.(((((...((((......))))...))))).)).....))))).)))).))))))...- ( -39.30, z-score = -0.88, R) >droAna3.scaffold_13337 13637606 103 - 23293914 GAUGCUAGGCGGCCAUAA-UUGUUUACUUUCAGGCAGUGCA-CGAGUCGCCCUGUGCUGGCUAAAAUGG---CAGCCCUGGUCAGCUGCUUUCG--GCCAGUGCUGCCAU----- ...(((((((((((...(-((((((......)))))))...-.).))))))......))))....((((---((((.((((((..........)--))))).))))))))----- ( -39.00, z-score = -0.40, R) >consensus _______UGUGACAAAAAAUUGUUUA_UAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUC_ .......((((((...................(((...((((....))))...)))(((((((((((((................)))).)))))))))......)))))).... (-26.15 = -26.82 + 0.67)
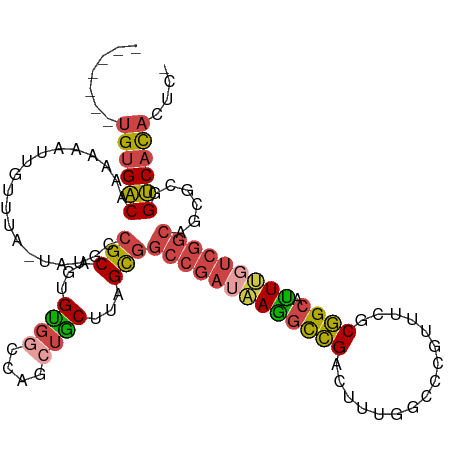
| Location | 3,787,992 – 3,788,105 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.22256 |
| G+C content | 0.58365 |
| Mean single sequence MFE | -51.24 |
| Consensus MFE | -37.00 |
| Energy contribution | -38.84 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3787992 113 + 24543557 AAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGAAUUUGCACCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG .............(((((.((((((((.(((((....(((((.....)))))................((((....)))))))))..))))))))((.....))..))))).. ( -49.10, z-score = -2.71, R) >droSim1.chr3L 3328044 113 + 22553184 AAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCACUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG .............(((((.((((((((.((((((....((((.....))))(((....(((((...)).)))....)))))))))..))))))))((.....))..))))).. ( -53.10, z-score = -3.16, R) >droSec1.super_2 3782047 113 + 7591821 AAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACAUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG .............(((((.((((((((.((((((....((((.....))))((((...(((((...)).)))...))))))))))..))))))))((.....))..))))).. ( -54.80, z-score = -3.97, R) >droYak2.chr3L 4343919 111 + 24197627 AAAUUGUUUAUAUGGCGCAGUGUGGCCAGCUACUAGGCGGCCGAUGAGGCCGAGUUUCGCCCGUUUGGCGGCAUUCUUCGGCAGCGCGGUCACAC-CGCGGCUGUA-UGAAUG .....(((((((((((((.((((((((.(((.((....((((.....))))((((.(((((.....))))).))))...)).)))..))))))))-.)).))))))-))))). ( -51.90, z-score = -3.04, R) >droEre2.scaffold_4784 6483589 112 + 25762168 AAAUUGUUUAUAUGGCGCAGUGUGGCCAGAUACUUGGCGGCCGAUAAGGCCGAGUUGGGCCCGUUUGGCGGCAUUUUUCGGCAGCGCGGUCACAC-CGCGGCUGUAGUGAGCG .....(((((((((((((.((((((((.((......((.(((((...((((......))))...))))).)).....)).((...))))))))))-.)).))))).)))))). ( -47.30, z-score = -1.53, R) >consensus AAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG .............((((((((((((((.((((((....((((.....))))(((....(.(((.....))))....)))))))))..)))))))))..........))))).. (-37.00 = -38.84 + 1.84)

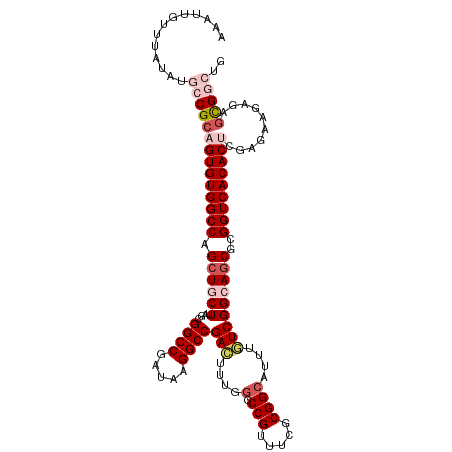
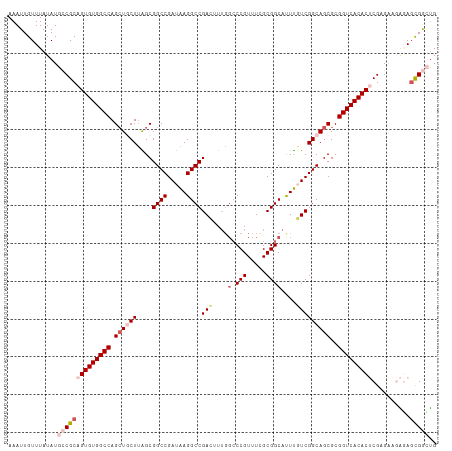
| Location | 3,787,992 – 3,788,105 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.22256 |
| G+C content | 0.58365 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -32.14 |
| Energy contribution | -32.02 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3787992 113 - 24543557 CAGCCGCUCUCUUCUCGAGUGUGACCGCGCUGCCGACAAAUGCCGCGAAACGGUGCAAAUUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUU ..(((((..........((((((.(((.(((((........((((.....)))).......(((((.....)))))....)))))))).)))))))))))............. ( -42.50, z-score = -2.40, R) >droSim1.chr3L 3328044 113 - 22553184 CAGCCGCUCUCUUCUCGAGUGUGACCGCGCUGCCGACAAGUGCCGCGAAACGGGCCAAAGUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUU ..(((((..........((((((.(((.(((((.(((....(((.((...)))))....)))((((.....)))).....)))))))).)))))))))))............. ( -44.70, z-score = -2.11, R) >droSec1.super_2 3782047 113 - 7591821 CAGCCGCUCUCUUCUCGAGUGUGACCGCGCUGCCGACAAAUGCCGCGAAACGGGCCAAUGUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUU ..(((((..........((((((.(((.(((((.((((...(((.((...)))))...))))((((.....)))).....)))))))).)))))))))))............. ( -46.40, z-score = -3.07, R) >droYak2.chr3L 4343919 111 - 24197627 CAUUCA-UACAGCCGCG-GUGUGACCGCGCUGCCGAAGAAUGCCGCCAAACGGGCGAAACUCGGCCUCAUCGGCCGCCUAGUAGCUGGCCACACUGCGCCAUAUAAACAAUUU ......-....(.((((-(((((.(((.(((((..........((((.....)))).....(((((.....)))))....)))))))).))))))))).)............. ( -42.40, z-score = -2.36, R) >droEre2.scaffold_4784 6483589 112 - 25762168 CGCUCACUACAGCCGCG-GUGUGACCGCGCUGCCGAAAAAUGCCGCCAAACGGGCCCAACUCGGCCUUAUCGGCCGCCAAGUAUCUGGCCACACUGCGCCAUAUAAACAAUUU .(((......)))((((-(((((.(((....(((((.....(((........))).....))))).....)))..((((......)))))))))))))............... ( -36.40, z-score = -0.97, R) >consensus CAGCCGCUCUCUUCUCGAGUGUGACCGCGCUGCCGACAAAUGCCGCGAAACGGGCCAAACUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUU ..............(((((((((.(((.(((((.........(((.....)))........(((((.....)))))....)))))))).)))))).))).............. (-32.14 = -32.02 + -0.12)

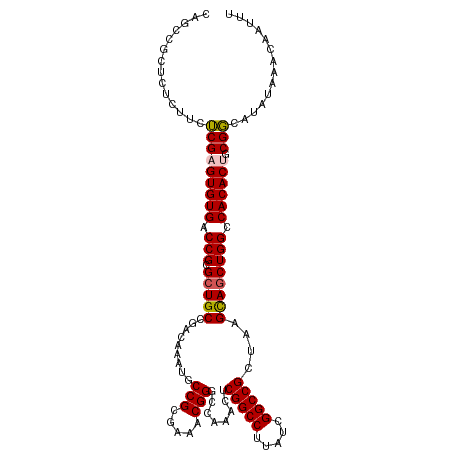
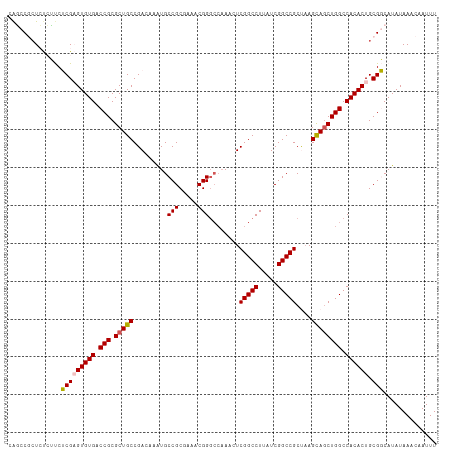
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:28 2011