
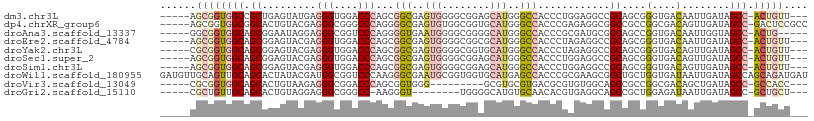
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,780,858 – 3,780,957 |
| Length | 99 |
| Max. P | 0.681363 |

| Location | 3,780,858 – 3,780,957 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.32 |
| Shannon entropy | 0.51228 |
| G+C content | 0.67594 |
| Mean single sequence MFE | -46.39 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
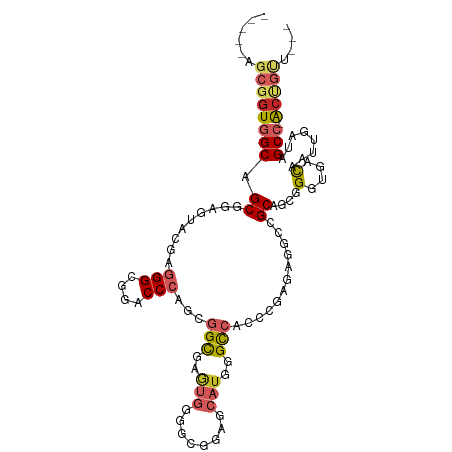
>dm3.chr3L 3780858 99 + 24543557 -----AGCGGUGGCCGCUGAGUAUGAGGGUGGACCCAGCGGCGAGUGGGGCGGAGCAUGGGCCACCCUGGAGGCCGCAGCGGGUGACAAUUGAUAGCC-ACUGUU--- -----((((((((((((((.((.(.(((((((.((((((..((.......))..)).))))))))))).)..))..)))))(....)........)))-))))))--- ( -48.90, z-score = -2.64, R) >dp4.chrXR_group6 6839244 102 + 13314419 -----AGCGGUGGCGGCACUGUACGAGGGCGGGCCCAGGGGCGAGUGUGGCGGUGCAUGGGCCACCCGAGAGGCGGCCGCCGGCGACAGUUGAUAGCC-GACUCCGCC -----.((((.(.(((((((((.((..((((((((((...(((.(.....)..))).)))))).(((....)).)..))))..))))))).....)))-).).)))). ( -49.80, z-score = -0.22, R) >droAna3.scaffold_13337 13630729 97 - 23293914 -----GGCGGUGGCAGCGGAAUAGGAGGGCGGUCCCAGGGGUGAAUGGGGCGGGGCAUGGGCCACCCGCGAUGCGGCAGCCGGUGACAAUUGGUAGCC-ACUG----- -----..(((((((.((.(....(..((((.((((((........)))))).)(((....))).))).)....).)).((((((....)))))).)))-))))----- ( -41.40, z-score = -0.62, R) >droEre2.scaffold_4784 6476182 99 + 25762168 -----AGCGGUGGCAGCGGAGUACGAGGGUGGACCCAGCGGCGAGUGGGGCGGCGCAUGGGCCACCCUAGAGGCCGCAGCGGGUGACAAUUGAUAGCC-ACUGUU--- -----(((((((((.((((..(...(((((((.(((((((.((.......)).))).)))))))))))..)..))))....(....)........)))-))))))--- ( -54.00, z-score = -4.49, R) >droYak2.chr3L 4336267 99 + 24197627 -----CGCGGUGGCAGCGGAGUACGAGGGUGGACCCAGCGGCGAGUGGGGCGGUGCAUGGGCCACCCUAGAGGCCGCAGCGGGUGACAGUUGAUAGCC-ACUGUU--- -----.((((((((.((((..(...(((((((.(((((((.((.......)).))).)))))))))))..)..))))(((.(....).)))....)))-))))).--- ( -53.20, z-score = -4.03, R) >droSec1.super_2 3774954 99 + 7591821 -----AGCGGUGGCAGCGGAGUACGAGGGUGGACCCAGCGGCGAGUGGGGCGGAGCAUGGGCCACCCUGGAGGCCGCAGCGGGUGACAGUUGAUAGCC-ACUGUU--- -----(((((((((.((((..(.(.(((((((.((((((..((.......))..)).))))))))))).))..))))(((.(....).)))....)))-))))))--- ( -52.10, z-score = -3.94, R) >droSim1.chr3L 3320902 99 + 22553184 -----AGCGGUGGCAGCGGAGUACGAGGGUGGACCCAGCGGCGAGUGGGGCGGAGCAUGGGCCACCCUGGAGGCCGCAGCGGGUGACAGUUGAUAGCC-ACUGUU--- -----(((((((((.((((..(.(.(((((((.((((((..((.......))..)).))))))))))).))..))))(((.(....).)))....)))-))))))--- ( -52.10, z-score = -3.94, R) >droWil1.scaffold_180955 1680942 108 + 2875958 GAUGUUGCAGUUGCAGCACUAUACGAUGGCGGUCCCAAGGGCGAAUGCGGUGGUGCAUGAGCCACCCGCGAAGCGGCUGCUGGUGAUAAUUGAUAGCCAGCAGAUGAU ..((((((....)))))).....((.(.(((((((...))))......((((((......))))))))).)..)).((((((((.((.....)).))))))))..... ( -39.30, z-score = -0.67, R) >droVir3.scaffold_13049 20978337 90 - 25233164 -----CGCGGUGGCAGCACUGUAAGAGGGCGGACCCAGCGGUGGG---------GCGUGCGUGACGCGUGUGGCAGCCGCCGGCGACAGCUGAUAGCC-GCCACC--- -----...((((((.((.........(((....))).(((((..(---------.((..((.....))..)).).)))))((((....))))...)).-))))))--- ( -40.80, z-score = 0.52, R) >droGri2.scaffold_15110 10555034 90 + 24565398 -----CGCUGUUGCAGCACUGUAGGAGGGCGGGCC-AAGGGU--------UGGGGCAUGUGCAACACGUGAGGCAGCCGCUGGAGAUAAUUGAUAGCC-GCUGCU--- -----((((((((((.((((((......))))(((-......--------...))).))))))))).))).((((((.((((..((...))..)))).-))))))--- ( -32.30, z-score = 0.23, R) >consensus _____AGCGGUGGCAGCGGAGUACGAGGGCGGACCCAGCGGCGAGUGGGGCGGAGCAUGGGCCACCCGAGAGGCCGCAGCGGGUGACAAUUGAUAGCC_ACUGUU___ ......((.(((((..(.........(((....)))...(((..(((........)))..)))......)..))))).)).(((...........))).......... (-17.86 = -18.37 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:26 2011