| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,733,520 – 3,733,613 |
| Length | 93 |
| Max. P | 0.977314 |

| Location | 3,733,520 – 3,733,613 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.48 |
| Shannon entropy | 0.60168 |
| G+C content | 0.52014 |
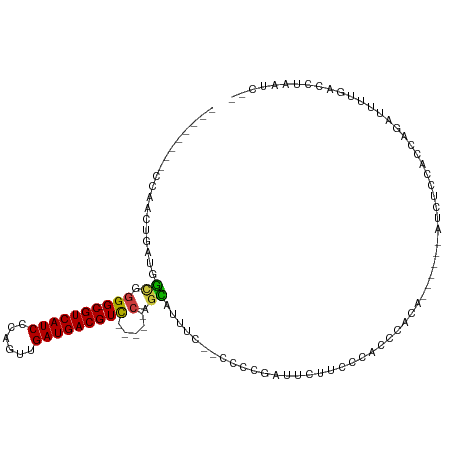
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.61 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
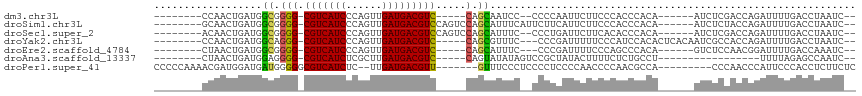
>dm3.chr3L 3733520 93 + 24543557 --------CCAACUGAUGGCGGGG-CGUCAUCCCAGUUGAUGACGUC-----CAGCAAUCC--CCCCAAUUCUUCCCACCCACA------AUCUCGACCAGAUUUUGACCUAAUC-- --------......(((.((.(((-(((((((......)))))))))-----).)).))).--.................((.(------((((.....))))).))........-- ( -24.30, z-score = -2.13, R) >droSim1.chr3L 3272554 100 + 22553184 --------GCAACUGAUGGCGGGG-CGUCAUCCCAGUUGAUGACGUCCAGUCCAGCAUUUCAUUCUUCAUUCUUCCCACCCACA------AUCUCUACCAGAUUUUGACCUAAUC-- --------....(((..(((.(((-(((((((......)))))))))).)))))).........................((.(------((((.....))))).))........-- ( -24.00, z-score = -1.93, R) >droSec1.super_2 3725043 98 + 7591821 --------ACAACUGAUGGCGGGG-CGUCAUCCCAGUUGAUGACGUCCAGUCCAGCAUUUC--CCCUGAUUCUUCACACCCACA------AUCUCGACCAGAUUUUGACCUAAUC-- --------....(((..(((.(((-(((((((......)))))))))).))))))......--...(((....)))....((.(------((((.....))))).))........-- ( -24.10, z-score = -1.05, R) >droYak2.chr3L 4286142 98 + 24197627 --------CCAACUGAUGGCAGGG-CGUCAUCCCAGUUGAUGACGUC-----CAGCGUUUC---CCCGAUUUUUCCCAUCCACACUCACAAUCGCCACCAGAUUUUGACCUAAUC-- --------....(((.((((.(((-(((((((......)))))))))-----)........---...((((..................)))))))).)))..............-- ( -27.47, z-score = -3.37, R) >droEre2.scaffold_4784 6429124 92 + 25762168 --------CUAACUGAUGGCGGGG-CGUCAUCCCAGUUGAUGACGUC-----CAGCAUUUC---CCCGAUUUUCCCAGCCCACA------GUCUCCAACGGAUUUUGACCAAAUC-- --------...((((..(((.(((-(((((((......)))))))))-----)........---...(.....)...)))..))------)).......((.......)).....-- ( -23.40, z-score = -0.69, R) >droAna3.scaffold_13337 13586466 84 - 23293914 --------CUAACUGAUGGAGGGG-CGUCAUCUCGCUUGAUGACGUC-----CAGUAUAUAGUCCGCUAUACUUUUCUCUGCCU-----------------UUUUAGAGCCAAUC-- --------((((..(..(((((((-(((((((......)))))))))-----)....(((((....))))).....))))..).-----------------..))))........-- ( -27.10, z-score = -2.95, R) >droPer1.super_41 138062 99 + 728894 CCCCCAAAACGAUGGAUGAUGGGGGCGUCAUCUC--UUGAUGACGUU-------GUUUCCCUCCCCUCCCCAACCCCAACGCCA---------CCCAACCCAUUCCCACCUCUUCUC ..........(((((....(((((((((((((..--..))))))(((-------(.....................))))))).---------))))..)))))............. ( -25.80, z-score = -2.94, R) >consensus ________CCAACUGAUGGCGGGG_CGUCAUCCCAGUUGAUGACGUC_____CAGCAUUUC__CCCCGAUUCUUCCCACCCACA______AUCUCCACCAGAUUUUGACCUAAUC__ .................((.((((.(((((((......)))))))............................)))).))..................................... (-10.68 = -10.61 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:22 2011