| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,725,048 – 3,725,098 |
| Length | 50 |
| Max. P | 0.979414 |

| Location | 3,725,048 – 3,725,098 |
|---|---|
| Length | 50 |
| Sequences | 8 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Shannon entropy | 0.42548 |
| G+C content | 0.44554 |
| Mean single sequence MFE | -8.48 |
| Consensus MFE | -6.79 |
| Energy contribution | -6.64 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3725048 50 + 24543557 CAAACUAUUCGAAACAGAUACG-------AAUUCCACUGGCAGAUUGCCAGC-CUCCG-- ......(((((.........))-------)))....(((((.....))))).-.....-- ( -9.30, z-score = -2.21, R) >droSim1.chr3L 3263984 50 + 22553184 CAAACUAUUCGAAACAGAUACG-------AAUUCCACUGGCAGAUUGCCAGC-CUCCG-- ......(((((.........))-------)))....(((((.....))))).-.....-- ( -9.30, z-score = -2.21, R) >droSec1.super_2 3716658 50 + 7591821 CAAACUAUUCGAAACAGAUACG-------AAUUCCACUGGCAGAUUGCCAGC-CUCCG-- ......(((((.........))-------)))....(((((.....))))).-.....-- ( -9.30, z-score = -2.21, R) >droYak2.chr3L 4277427 50 + 24197627 CAAACUAUUCGAAACAGAUACG-------AAUUCCGCUGGCAAAAUGCCAGC-CUUCG-- ......(((((.........))-------)))...((((((.....))))))-.....-- ( -12.90, z-score = -3.55, R) >droEre2.scaffold_4784 6420359 50 + 25762168 CAAACUAUUCGAAACAGAUACG-------AAUUCCGCUGGCAAAUUGCCAAC-CUUCG-- ......(((((.........))-------))).....((((.....))))..-.....-- ( -6.70, z-score = -1.33, R) >dp4.chrXR_group6 6767348 50 + 13314419 AUGCCUAUUUGAAACAGAUACA-------AAUUCCGUUGGCCCAAUGCCAGA-CAGAG-- .....((((((...))))))..-------...((.((((((.....)))).)-).)).-- ( -6.60, z-score = -0.60, R) >droWil1.scaffold_180955 1604302 56 + 2875958 UAAACUAUUUGAAACAAAUACAUAGUACAAAUUCUAUUGGCUCUUGGCCAAAACUG---- .....((((((...))))))................(((((.....))))).....---- ( -7.20, z-score = -1.74, R) >droMoj3.scaffold_6680 22015545 52 - 24764193 -AAACUAUUUGAAACAGAUACA-------AAUUCCGUUGGCCCAGAGCCAGAACGCCGCC -....((((((...))))))..-------.....(((((((.....))))..)))..... ( -6.50, z-score = -0.39, R) >consensus CAAACUAUUCGAAACAGAUACG_______AAUUCCACUGGCACAUUGCCAGC_CUCCG__ ....................................(((((.....)))))......... ( -6.79 = -6.64 + -0.16)
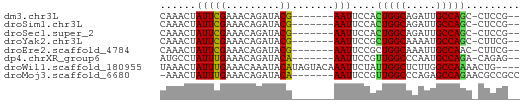
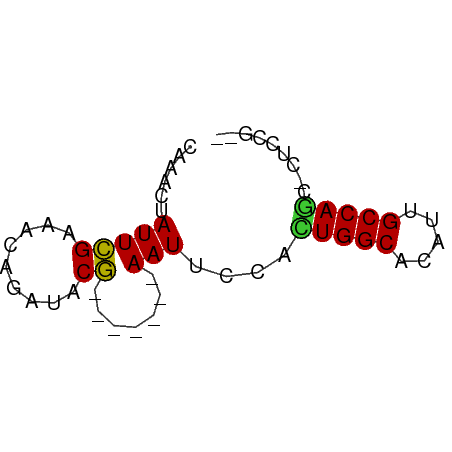
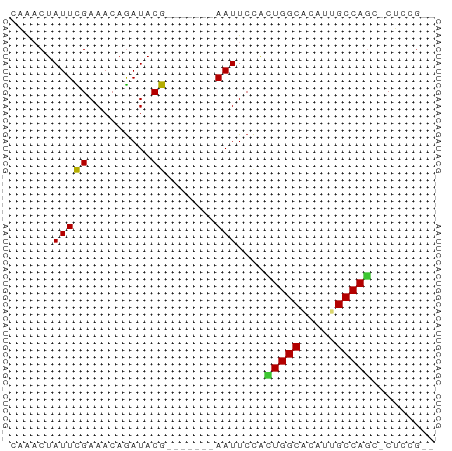
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:21 2011