
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,718,520 – 3,718,631 |
| Length | 111 |
| Max. P | 0.997701 |

| Location | 3,718,520 – 3,718,631 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 64.81 |
| Shannon entropy | 0.60944 |
| G+C content | 0.37099 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -8.87 |
| Energy contribution | -11.07 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
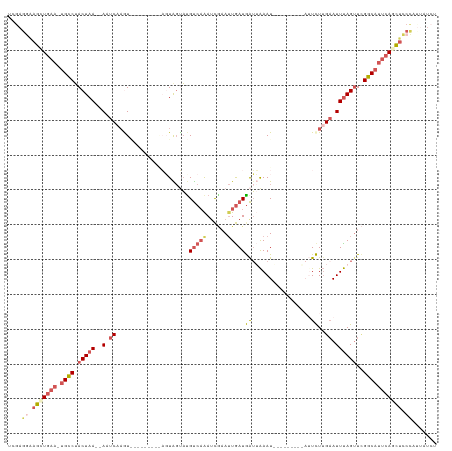
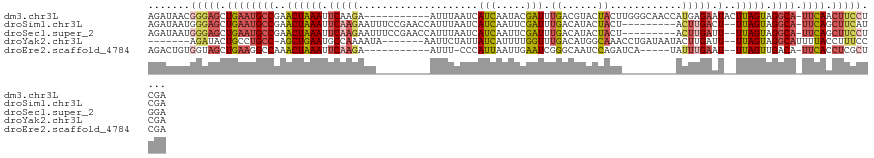
>dm3.chr3L 3718520 111 + 24543557 UCGAGGAAGUUGAA-UGCCUACUAAGUAUUCUCAUGGUUGCCCAAGUAGUACGUCAAAUCGUAUUGAUGAUUAAAU-----------UCUUGAAUUUAGUUCGGCAUUCAGCUCCCGUUAUCU .((.(((.((((((-((((.(((((((....(((.((((.......(((((((......))))))).......)))-----------)..))))))))))..)))))))))))))))...... ( -33.84, z-score = -4.36, R) >droSim1.chr3L 3257610 111 + 22553184 UCGAUGAAGCUGAA-UGCCUACUAA--AGUCAAGU---------AGUAGUAUGUCAAAUCGAAUUGAUGAUUAAAUGGUUCGGAAAUUCUUGAAUUUAGUUCGGCAUUCAGCUCCCAUUAUCU ..((((((((((((-((((.(((((--(.(((((.---------(((.(.....)...(((((((.((......)))))))))..)))))))).))))))..)))))))))))....))))). ( -34.60, z-score = -4.48, R) >droSec1.super_2 3710342 111 + 7591821 UCCAGGAAGCUGAA-UGCCUACUAA--AAUCAAGU---------AGUAGUAUGUCAAAUCGAAUUGAUGAUUAAAUGGUUCGGAAAUUCUUGAAUUUAGUUCGGCAUUCAGCUCCCAUUAUCU ....((.(((((((-((((.(((((--(.(((((.---------(((.(.....)...(((((((.((......)))))))))..)))))))).))))))..))))))))))).))....... ( -36.00, z-score = -5.04, R) >droYak2.chr3L 4270861 106 + 24197627 UCGGGAAAGGUAAAAUGCCUACUAA--AAUCAAGUAUUAUCAGGUUUGCCAUGUCAAACCAAAAUGAUAAUAGAAUU-------UAUUUUGGCAUUCAGCU-GGCAGGCAGUAUCU------- .......(((((...(((((.....--.......((((((((((((((......))))))....)))))))).....-------....(..((.....)).-.).))))).)))))------- ( -25.10, z-score = -1.66, R) >droEre2.scaffold_4784 6413975 103 + 25762168 UCGAGCGAGGUGAA-UGUCAACUAA--AUUCAAAUA-----UGAUCUGGAUUGCCCGAUUCAAUUAAUGGG-AAAU-----------UCUUGAAUUUAGUUUGGCCUUCAGCUACCACAGUCU .......((.((((-.(((((((((--((((((...-----......(((((.((((.(......).))))-.)))-----------)))))))))))).))))).)))).)).......... ( -26.10, z-score = -1.56, R) >consensus UCGAGGAAGCUGAA_UGCCUACUAA__AAUCAAGU_________AGUAGUAUGUCAAAUCGAAUUGAUGAUUAAAU_________AUUCUUGAAUUUAGUUCGGCAUUCAGCUCCCAUUAUCU ....((.(((((((.((((................................((((((......))))))(((((((.................)))))))..))))))))))).))....... ( -8.87 = -11.07 + 2.20)


| Location | 3,718,520 – 3,718,631 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 64.81 |
| Shannon entropy | 0.60944 |
| G+C content | 0.37099 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -7.44 |
| Energy contribution | -9.04 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
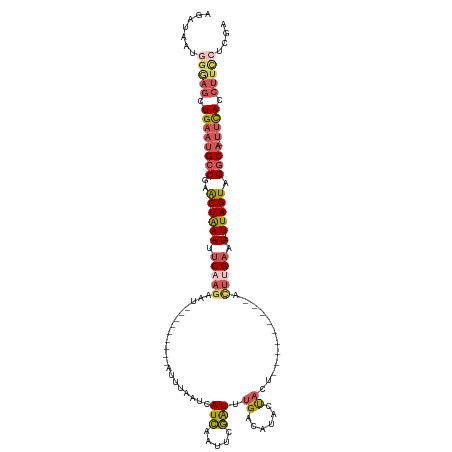
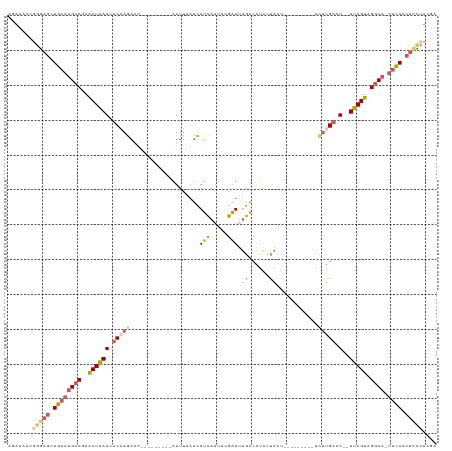
>dm3.chr3L 3718520 111 - 24543557 AGAUAACGGGAGCUGAAUGCCGAACUAAAUUCAAGA-----------AUUUAAUCAUCAAUACGAUUUGACGUACUACUUGGGCAACCAUGAGAAUACUUAGUAGGCA-UUCAACUUCCUCGA .......(((((.((((((((..(((((((((..((-----------......)).(((.((((......)))).....(((....))))))))))..))))).))))-)))).))))).... ( -33.70, z-score = -4.89, R) >droSim1.chr3L 3257610 111 - 22553184 AGAUAAUGGGAGCUGAAUGCCGAACUAAAUUCAAGAAUUUCCGAACCAUUUAAUCAUCAAUUCGAUUUGACAUACUACU---------ACUUGACU--UUAGUAGGCA-UUCAGCUUCAUCGA ........(((((((((((((..((((((.(((((.......((.........)).((((......)))).........---------.))))).)--))))).))))-)))))))))..... ( -35.50, z-score = -5.98, R) >droSec1.super_2 3710342 111 - 7591821 AGAUAAUGGGAGCUGAAUGCCGAACUAAAUUCAAGAAUUUCCGAACCAUUUAAUCAUCAAUUCGAUUUGACAUACUACU---------ACUUGAUU--UUAGUAGGCA-UUCAGCUUCCUGGA .......((((((((((((((..((((((.(((((.......((.........)).((((......)))).........---------.))))).)--))))).))))-)))))))))).... ( -38.80, z-score = -6.55, R) >droYak2.chr3L 4270861 106 - 24197627 -------AGAUACUGCCUGCC-AGCUGAAUGCCAAAAUA-------AAUUCUAUUAUCAUUUUGGUUUGACAUGGCAAACCUGAUAAUACUUGAUU--UUAGUAGGCAUUUUACCUUUCCCGA -------......(((((((.-.((.....)).....((-------((.((((((((((....((((((......))))))))))))))...)).)--))))))))))............... ( -24.40, z-score = -2.48, R) >droEre2.scaffold_4784 6413975 103 - 25762168 AGACUGUGGUAGCUGAAGGCCAAACUAAAUUCAAGA-----------AUUU-CCCAUUAAUUGAAUCGGGCAAUCCAGAUCA-----UAUUUGAAU--UUAGUUGACA-UUCACCUCGCUCGA .((.((.(((....(((.....(((((((((((((.-----------....-..((.....)).(((.((....)).)))..-----..)))))))--))))))....-)))))).)).)).. ( -19.40, z-score = 0.11, R) >consensus AGAUAAUGGGAGCUGAAUGCCGAACUAAAUUCAAGAAU_________AUUUAAUCAUCAAUUCGAUUUGACAUACUACU_________ACUUGAAU__UUAGUAGGCA_UUCACCUUCCUCGA .......(((((.((((((((..(((((..(((((....................(((.....))).((......))............)))))....))))).)))).)))).))))).... ( -7.44 = -9.04 + 1.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:20 2011