| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,711,114 – 3,711,212 |
| Length | 98 |
| Max. P | 0.640095 |

| Location | 3,711,114 – 3,711,212 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.11 |
| Shannon entropy | 0.52710 |
| G+C content | 0.43858 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -13.39 |
| Energy contribution | -14.50 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
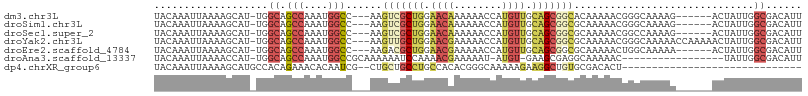
>dm3.chr3L 3711114 98 + 24543557 UACAAAUUAAAAGCAU-UGGCAGCCAAAUGGCC---AAGUCGCUGGAACAAAAAACCAUGUUGCAGCGGCACAAAAACGGGCAAAAG------ACUAUUGGCGACAUU ............((..-..)).(((((...(((---..(((((((.((((........)))).))))))).(......)))).....------....)))))...... ( -25.90, z-score = -1.63, R) >droSim1.chr3L 3250275 98 + 22553184 UACAAAUUAAAAGCAU-UGGCAGCCAAAUGGCC---AAGUCGCUGGAACAAAAAACCAUGUUGCAGCGGCGCAAAAACGGGCAAAAG------ACUAUUGGCGACAUU ............((..-..)).(((((.(((.(---..(((((((.((((........)))).)))))))((........))....)------.))))))))...... ( -28.00, z-score = -1.83, R) >droSec1.super_2 3703063 98 + 7591821 UACAAAUUAAAAGCAU-UGGCAGCCAAAUGGCC---AAGUCGCUGGAACAAAAAACCAUGUUGCAGCGGCGCAAAAACGGCCAAAAG------ACUAUUGGCGACAUU ............((.(-((((.........)))---))(((((((.((((........)))).))))))))).......(((((...------....)))))...... ( -30.60, z-score = -2.66, R) >droYak2.chr3L 4263256 104 + 24197627 UACAAAUUAAAAGCAU-UGGCAGCCAAAUGGCC---AAGUUGCUGGAACGAAAAACCAUGUUGCAGCGGCGCAAAAACGGGCAAAAACCAAAAACUAUUGGCGACAUU ............((..-..)).(((((.(((..---..(((((((.((((........)))).)))))))........((.......)).....))))))))...... ( -24.80, z-score = -0.32, R) >droEre2.scaffold_4784 6406680 98 + 25762168 UACAAAUUAAAAGCAU-UGGCAGCCAAAUGGCC---AAGACGCUGGAACGAAAAACCAUGUUGCAGCGGCGCAAAAACUGGCAAAAA------ACUAUUGGCGACAUU ............((..-..)).(((((.(((..---....(((((.((((........)))).)))))((.((.....)))).....------.))))))))...... ( -22.50, z-score = -0.36, R) >droAna3.scaffold_13337 13564272 88 - 23293914 UACAAAUUAAAACCAU-UGGCAGCCAAAUGGCCGCAAAAAAUCCAAAACGAAAAAU-AUGU-GAAGCGAGGCAAAAAC-----------------UAUUGGCGACAUU ................-..((.(((....))).)).....................-..((-(..((.((........-----------------..)).))..))). ( -10.50, z-score = 1.16, R) >dp4.chrXR_group6 6753063 76 + 13314419 UACAAAUUAAAAGCAUGCCACAGAAACACAAUCG--CUGCUGCCUGCCACACGGGCAAAAAGAAGGCUGUGCGACACU------------------------------ ............(((((((.(((...........--))).((((((.....)))))).......))).))))......------------------------------ ( -18.40, z-score = -1.01, R) >consensus UACAAAUUAAAAGCAU_UGGCAGCCAAAUGGCC___AAGUCGCUGGAACAAAAAACCAUGUUGCAGCGGCGCAAAAACGGGCAAAAG______ACUAUUGGCGACAUU ...................((.(((....)))......(((((((.((((........)))).)))))))..............................))...... (-13.39 = -14.50 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:18 2011