
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 464,856 – 464,950 |
| Length | 94 |
| Max. P | 0.790313 |

| Location | 464,856 – 464,950 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.17 |
| Shannon entropy | 0.44663 |
| G+C content | 0.59762 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -20.50 |
| Energy contribution | -22.31 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.786112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
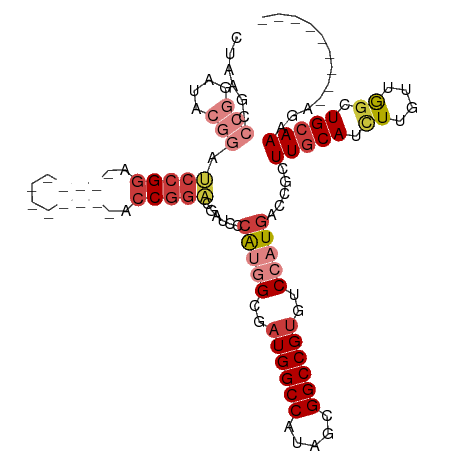
>dm3.chr2L 464856 94 + 23011544 CUAAGCCCGGAUACGGAUCCGGA------------ACCGGACGAUCCCAUGGCAAUGGCCAUAGCGGCCGUGUCCAUGACCGCUUGCAUCUUGUUGGCUGCAAAGA--------- ..((((..((((.((..((((..------------..))))))))))(((((..((((((.....))))))..)))))...))))(((.((....)).))).....--------- ( -34.50, z-score = -1.22, R) >droSim1.chr2L 471425 94 + 22036055 CUAAGCCCGGAUACGGAUCCGGA------------ACCGGACGAUCCCAUGGCGAUGGCCAUAGCGGCCGUGUCCAUGACCGCUUGCAUCUUGUUGGCUGCAAAGA--------- ..((((..((((.((..((((..------------..))))))))))(((((..((((((.....))))))..)))))...))))(((.((....)).))).....--------- ( -34.50, z-score = -1.03, R) >droSec1.super_14 448024 94 + 2068291 CUAAGCCCGGAUACGGAUCCGGA------------ACCGGACGAUCCCAUGGCGAUGGCCAUAGCGGCCGUGUCCAUGACCGCUUGCAUCUUGUUGGCUGCAAAGA--------- ..((((..((((.((..((((..------------..))))))))))(((((..((((((.....))))))..)))))...))))(((.((....)).))).....--------- ( -34.50, z-score = -1.03, R) >droYak2.chr2L 449511 100 + 22324452 CUAAGCCCGGAUACGGAUCCGGAGCC------GGAACCGGACGACCCCAUGGCGAUGGCCAUGGCGGCCGUGUCCAUGACGGCUUGCAUCUUGUUGGCUGCAAAGA--------- ........((((((((.(((((....------....)))))...(((((((((....))))))).)))))))))).((.(((((.((.....)).)))))))....--------- ( -45.20, z-score = -2.42, R) >droEre2.scaffold_4929 516806 106 + 26641161 CUAAGCCCGGAUACGGAUCCGGAGCCGGAGCCGGAACCGGACGAUCCCAUGGCGAUGGCCAUAGCGGCCGUGUCCAUGACCGCUUGCAUCUUGUUGACUGCAAAGA--------- ..((((..((((.((..(((((..(((....)))..)))))))))))(((((..((((((.....))))))..)))))...))))(((((.....)).))).....--------- ( -43.30, z-score = -2.13, R) >droAna3.scaffold_12943 257348 91 - 5039921 ------------ACCGAGCCGGA------------UCCGGGUGCACCCACAGAGAUGGCCAUGGCGGCCGUGUCCAUGGCCGCUUGCAUUUUUCUGGCUGGAAAUGGAAAAUUAA ------------.(((..((((.------------...(((....))).((((((..((...(((((((((....))))))))).))...)))))).))))...)))........ ( -38.90, z-score = -2.22, R) >droWil1.scaffold_180772 5946513 76 - 8906247 ---------------GAGCUGCU------------GCCCAUGGCUGCAGCAGCAAUGGCCAUGGAGGCCGAGUCCAUGACCGCUUGCAUCUUGUAAGCUGCAA------------ ---------------..(((((.------------(((...))).))))).(((..((.((((((.......)))))).))((((((.....)))))))))..------------ ( -34.20, z-score = -1.82, R) >consensus CUAAGCCCGGAUACGGAUCCGGA____________ACCGGACGAUCCCAUGGCGAUGGCCAUAGCGGCCGUGUCCAUGACCGCUUGCAUCUUGUUGGCUGCAAAGA_________ ......(((....))).(((((..............)))))......(((((..((((((.....))))))..))))).....(((((.((....)).)))))............ (-20.50 = -22.31 + 1.81)

| Location | 464,856 – 464,950 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.17 |
| Shannon entropy | 0.44663 |
| G+C content | 0.59762 |
| Mean single sequence MFE | -37.51 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.91 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
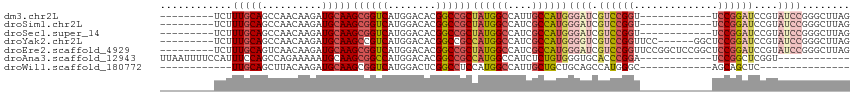
>dm3.chr2L 464856 94 - 23011544 ---------UCUUUGCAGCCAACAAGAUGCAAGCGGUCAUGGACACGGCCGCUAUGGCCAUUGCCAUGGGAUCGUCCGGU------------UCCGGAUCCGUAUCCGGGCUUAG ---------.......((((.....(((((.(((((((........)))))))(((((....))))).(((...((((..------------..))))))))))))..))))... ( -35.80, z-score = -1.32, R) >droSim1.chr2L 471425 94 - 22036055 ---------UCUUUGCAGCCAACAAGAUGCAAGCGGUCAUGGACACGGCCGCUAUGGCCAUCGCCAUGGGAUCGUCCGGU------------UCCGGAUCCGUAUCCGGGCUUAG ---------.......((((.....(((((.(((((((........)))))))(((((....))))).(((...((((..------------..))))))))))))..))))... ( -35.90, z-score = -1.30, R) >droSec1.super_14 448024 94 - 2068291 ---------UCUUUGCAGCCAACAAGAUGCAAGCGGUCAUGGACACGGCCGCUAUGGCCAUCGCCAUGGGAUCGUCCGGU------------UCCGGAUCCGUAUCCGGGCUUAG ---------.......((((.....(((((.(((((((........)))))))(((((....))))).(((...((((..------------..))))))))))))..))))... ( -35.90, z-score = -1.30, R) >droYak2.chr2L 449511 100 - 22324452 ---------UCUUUGCAGCCAACAAGAUGCAAGCCGUCAUGGACACGGCCGCCAUGGCCAUCGCCAUGGGGUCGUCCGGUUCC------GGCUCCGGAUCCGUAUCCGGGCUUAG ---------.......((((.....(((((..........((((..((((.(((((((....)))))))))))))))((.(((------(....)))).)))))))..))))... ( -44.40, z-score = -2.46, R) >droEre2.scaffold_4929 516806 106 - 26641161 ---------UCUUUGCAGUCAACAAGAUGCAAGCGGUCAUGGACACGGCCGCUAUGGCCAUCGCCAUGGGAUCGUCCGGUUCCGGCUCCGGCUCCGGAUCCGUAUCCGGGCUUAG ---------...(((((.((.....)))))))((((((........))))))((((((....)))))).....((((((...(((.((((....)))).)))...)))))).... ( -44.80, z-score = -2.14, R) >droAna3.scaffold_12943 257348 91 + 5039921 UUAAUUUUCCAUUUCCAGCCAGAAAAAUGCAAGCGGCCAUGGACACGGCCGCCAUGGCCAUCUCUGUGGGUGCACCCGGA------------UCCGGCUCGGU------------ ..............((((((.((...........((((((((.(......)))))))))...((((.((.....))))))------------)).)))).)).------------ ( -31.10, z-score = -0.50, R) >droWil1.scaffold_180772 5946513 76 + 8906247 ------------UUGCAGCUUACAAGAUGCAAGCGGUCAUGGACUCGGCCUCCAUGGCCAUUGCUGCUGCAGCCAUGGGC------------AGCAGCUC--------------- ------------(((((.((....)).)))))..(((((((((.......)))))))))...((((((((........))------------))))))..--------------- ( -34.70, z-score = -2.10, R) >consensus _________UCUUUGCAGCCAACAAGAUGCAAGCGGUCAUGGACACGGCCGCUAUGGCCAUCGCCAUGGGAUCGUCCGGU____________UCCGGAUCCGUAUCCGGGCUUAG ............(((((..........)))))((((((........))))))((((((....))))))(((((...(((..............)))))))).............. (-21.40 = -21.91 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:56 2011