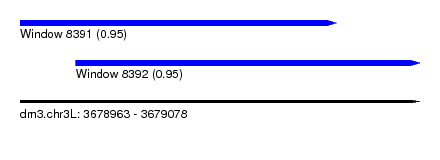
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,678,963 – 3,679,078 |
| Length | 115 |
| Max. P | 0.952778 |

| Location | 3,678,963 – 3,679,054 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 86.91 |
| Shannon entropy | 0.23831 |
| G+C content | 0.40467 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.49 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3678963 91 + 24543557 -------------------------------ACGCCGUCCUCAAUUGCCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -------------------------------..(((((..(((..........))))))))(((((.((((((......(((((((((....))))))))).)))))))))))......... ( -24.50, z-score = -2.98, R) >droSim1.chr3L 3217101 91 + 22553184 -------------------------------ACGCCGUCCUCAAUUGCCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -------------------------------..(((((..(((..........))))))))(((((.((((((......(((((((((....))))))))).)))))))))))......... ( -24.50, z-score = -2.98, R) >droSec1.super_2 3670647 91 + 7591821 -------------------------------ACGCCGUCCUCAAUUGCCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -------------------------------..(((((..(((..........))))))))(((((.((((((......(((((((((....))))))))).)))))))))))......... ( -24.50, z-score = -2.98, R) >droYak2.chr3L 4228701 91 + 24197627 -------------------------------ACGCCGUCCUCAAUUGCCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -------------------------------..(((((..(((..........))))))))(((((.((((((......(((((((((....))))))))).)))))))))))......... ( -24.50, z-score = -2.98, R) >droEre2.scaffold_4784 6373620 91 + 25762168 -------------------------------ACGCCGUCCUCAAUUGCGCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -------------------------------.....((((......(((((.......)))))...(((((((......(((((((((....))))))))).)))))))))))......... ( -24.60, z-score = -2.40, R) >droAna3.scaffold_13337 13528896 91 - 23293914 -------------------------------UCGCCGUCCUCAAUUGCCUCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -------------------------------..(((((..(((..........))))))))(((((.((((((......(((((((((....))))))))).)))))))))))......... ( -24.50, z-score = -2.97, R) >droPer1.super_41 65067 120 + 728894 GCGUCUGCGUCCGCGUCUGCGUCCUCAAUUGGAGUCUCUCUGAAUGG--UCUCUGAAUGGCGUCCGAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA (((..(((....)))..)))((((((((((((((((..((.((....--..)).))..))).))))))).))..((((.(((((((((....)))))))))....))))))))......... ( -33.30, z-score = -1.41, R) >droWil1.scaffold_180955 1533838 90 + 2875958 -------------------------------CUGUCUCUGUGAGUCGC-UCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -------------------------------..(((.....(((....-.))).....)))(((((.((((((......(((((((((....))))))))).)))))))))))......... ( -23.30, z-score = -2.23, R) >droVir3.scaffold_13049 20831444 87 - 25233164 -----------------------------------CGUCCUCAAUUGCCUGGCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGCUGAAUGGACAAUUAUGAA -----------------------------------.....(((...(((.........)))(((((.(((((....)).(((((((((....)))))))))....)))))))).....))). ( -21.50, z-score = -1.59, R) >droMoj3.scaffold_6680 21948192 87 - 24764193 -----------------------------------CGUCCUCAAUUGCCUGGCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -----------------------------------.....(((...(((.........)))(((((.((((((......(((((((((....))))))))).))))))))))).....))). ( -23.20, z-score = -2.24, R) >droGri2.scaffold_15110 10435673 87 + 24565398 -----------------------------------CGUCCUCAAUUGCCUGGCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA -----------------------------------.....(((...(((.........)))(((((.((((((......(((((((((....))))))))).))))))))))).....))). ( -23.20, z-score = -2.24, R) >consensus _______________________________ACGCCGUCCUCAAUUGCCUCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAA ..............................................((...........))(((((.((((((......(((((((((....))))))))).)))))))))))......... (-18.39 = -18.49 + 0.10)

| Location | 3,678,979 – 3,679,078 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.21 |
| Shannon entropy | 0.18414 |
| G+C content | 0.34312 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.71 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3678979 99 + 24543557 -----CCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUCU--GAACA-------------- -----..((.......)).(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))(((((.....)--)))).-------------- ( -23.30, z-score = -2.28, R) >droSim1.chr3L 3217117 99 + 22553184 -----CCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUCU--GAACA-------------- -----..((.......)).(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))(((((.....)--)))).-------------- ( -23.30, z-score = -2.28, R) >droSec1.super_2 3670663 99 + 7591821 -----CCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUCU--GAACA-------------- -----..((.......)).(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))(((((.....)--)))).-------------- ( -23.30, z-score = -2.28, R) >droYak2.chr3L 4228717 99 + 24197627 -----CCCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUCU--GAGCA-------------- -----...(((.(((....(((((.((((((......(((((((((....))))))))).))))))))))).....(((((.......)))))..))).--)))..-------------- ( -24.40, z-score = -2.47, R) >droEre2.scaffold_4784 6373636 99 + 25762168 -----CGCCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUCU--GAACA-------------- -----.(((.......)))(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))(((((.....)--)))).-------------- ( -28.00, z-score = -3.58, R) >droAna3.scaffold_13337 13528912 99 - 23293914 -----CCUCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUUU--GAACA-------------- -----..............(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))(((((.....)--)))).-------------- ( -23.00, z-score = -2.12, R) >dp4.chrXR_group6 6707836 115 + 13314419 UGAAUGGUCUCUGAAUGGCGUCCGAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUUUUCGACUACAGAGCACA----- ......(((((((......((((.(((((((......(((((((((....))))))))).))))))))))).....(((((.......))))).............))))).)).----- ( -28.80, z-score = -1.51, R) >droPer1.super_41 65107 120 + 728894 UGAAUGGUCUCUGAAUGGCGUCCGAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUUUUCGAGUACAGAGCACAGAGCA ......(.(((((....((((((.(((((((......(((((((((....))))))))).))))))))))).....(((((.......))))).................)).)))))). ( -29.10, z-score = -0.87, R) >droWil1.scaffold_180955 1533854 97 + 2875958 ------CUCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUC---GAACA-------------- ------.............(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))((((......---)))).-------------- ( -22.30, z-score = -1.75, R) >droVir3.scaffold_13049 20831456 97 - 25233164 -----CCUGGCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGCUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUU----GAACA-------------- -----....(((....)))(((((.(((((....)).(((((((((....)))))))))....))))))))....((((.....))))((((.....----)))).-------------- ( -23.00, z-score = -1.67, R) >droMoj3.scaffold_6680 21948204 97 - 24764193 -----CCUGGCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUU----GAACA-------------- -----....(((....)))(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))((((.....----)))).-------------- ( -24.70, z-score = -2.19, R) >droGri2.scaffold_15110 10435685 97 + 24565398 -----CCUGGCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUU----GAACA-------------- -----....(((....)))(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....))))((((.....----)))).-------------- ( -24.70, z-score = -2.19, R) >consensus _____CCUCUCUGAAUGGCGUCCAAUUCGAUUAUUCUAACAUGAAUUAUAGUUCGUGUUCGUUGAAUGGACAAUUAUGAAUUUAUCAUGUUCAAUUUCU__GAACA______________ ...........(((((...(((((.((((((......(((((((((....))))))))).)))))))))))....((((.....)))))))))........................... (-21.76 = -21.71 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:15 2011