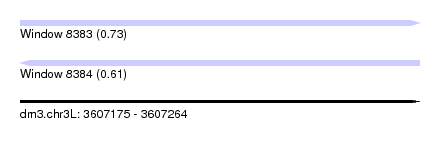
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,607,175 – 3,607,264 |
| Length | 89 |
| Max. P | 0.727548 |

| Location | 3,607,175 – 3,607,264 |
|---|---|
| Length | 89 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Shannon entropy | 0.41146 |
| G+C content | 0.37391 |
| Mean single sequence MFE | -15.61 |
| Consensus MFE | -8.80 |
| Energy contribution | -9.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
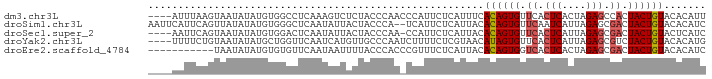
>dm3.chr3L 3607175 89 + 24543557 ----AUUUAAGUAAUAUAUGUGGCCUCAAAGUCUCUACCCAACCCAUUCUCAUUUCACAGUGUUCACUCACUAGAGCCACUACUGUACACAUU ----......(((......(((((.((...((....))....................((((......)))).))))))).....)))..... ( -13.00, z-score = -1.34, R) >droSim1.chr3L 3144647 91 + 22553184 AAUUCAUUCAGUUAUAUAUGUGGGCUCAAUAUUACUACCCA--UCAUUCUCAUUACACAGUGUUCAAUCAUUAGAGCGACUACUGUACACAUC ..........((.....(((((((.............))))--.))).......))((((((.((..((....))..)).))))))....... ( -13.02, z-score = -0.74, R) >droSec1.super_2 3593096 88 + 7591821 ----AAUUCAGUAAUAUAUGUGGACUCAAUAUUACUACCCAA-CCAUUCUCAUUACACAGUGUUCACUCAUUAGAGCGACUACUGUACUCAUC ----.....((((((((.((......))))))))))......-.............((((((.((.(((....))).)).))))))....... ( -17.40, z-score = -2.51, R) >droYak2.chr3L 4155091 89 + 24197627 ----UUUUCUGUAAUAUAUGCUGGUUCAAUCAUGUUGCCCAAUCUUUUCUCGUAACAUAGUGUUCACUCAUUAGAGCGUCUACUGUACACAUG ----............((((.((..........(((((.............)))))((((((..(.(((....))).)..)))))).)))))) ( -12.32, z-score = 0.04, R) >droEre2.scaffold_4784 6300922 82 + 25762168 -----------UAAUAUAUGUGUGUUCAAUAAUUUUACCCACCCGUUUCUCAUUACACAGUGGUCACUCACUAGAGCGACUACUGUACACAUC -----------......((((((.....................((........))(((((((((.(((....))).))))))))))))))). ( -22.30, z-score = -4.02, R) >consensus ____AAUUCAGUAAUAUAUGUGGGCUCAAUAUUACUACCCAA_CCAUUCUCAUUACACAGUGUUCACUCAUUAGAGCGACUACUGUACACAUC ........................................................((((((.((.(((....))).)).))))))....... ( -8.80 = -9.24 + 0.44)
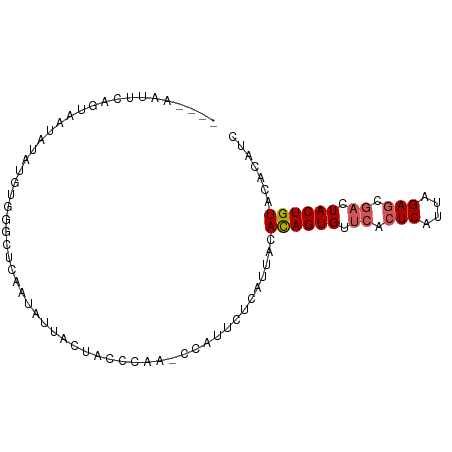

| Location | 3,607,175 – 3,607,264 |
|---|---|
| Length | 89 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 77.02 |
| Shannon entropy | 0.41146 |
| G+C content | 0.37391 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -12.76 |
| Energy contribution | -14.16 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
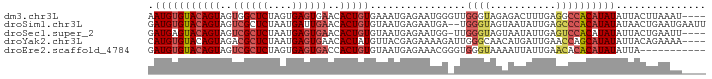
>dm3.chr3L 3607175 89 - 24543557 AAUGUGUACAGUAGUGGCUCUAGUGAGUGAACACUGUGAAAUGAGAAUGGGUUGGGUAGAGACUUUGAGGCCACAUAUAUUACUUAAAU---- .(((((((((((..(.((((....)))).)..)))))............((((.(((....)))....))))))))))...........---- ( -19.20, z-score = -0.50, R) >droSim1.chr3L 3144647 91 - 22553184 GAUGUGUACAGUAGUCGCUCUAAUGAUUGAACACUGUGUAAUGAGAAUGA--UGGGUAGUAAUAUUGAGCCCACAUAUAUAACUGAAUGAAUU .(((((((((((..(((.((....)).)))..))))).............--.((((...........))))))))))............... ( -17.40, z-score = -0.53, R) >droSec1.super_2 3593096 88 - 7591821 GAUGAGUACAGUAGUCGCUCUAAUGAGUGAACACUGUGUAAUGAGAAUGG-UUGGGUAGUAAUAUUGAGUCCACAUAUAUUACUGAAUU---- .......(((((..((((((....))))))..)))))((((((....(((-((.((((....)))).)).)))....))))))......---- ( -24.00, z-score = -2.31, R) >droYak2.chr3L 4155091 89 - 24197627 CAUGUGUACAGUAGACGCUCUAAUGAGUGAACACUAUGUUACGAGAAAAGAUUGGGCAACAUGAUUGAACCAGCAUAUAUUACAGAAAA---- ((((((..((((.....(((((((((((....))).))))).))).....))))..).)))))..........................---- ( -15.40, z-score = -0.45, R) >droEre2.scaffold_4784 6300922 82 - 25762168 GAUGUGUACAGUAGUCGCUCUAGUGAGUGACCACUGUGUAAUGAGAAACGGGUGGGUAAAAUUAUUGAACACACAUAUAUUA----------- .(((((((((((.(((((((....))))))).)))))..............(((..(((.....)))..)))))))))....----------- ( -25.30, z-score = -2.87, R) >consensus GAUGUGUACAGUAGUCGCUCUAAUGAGUGAACACUGUGUAAUGAGAAUGG_UUGGGUAGAAAUAUUGAGCCCACAUAUAUUACUGAAAU____ .(((((((((((..((((((....))))))..)))))................((((...........))))))))))............... (-12.76 = -14.16 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:08 2011