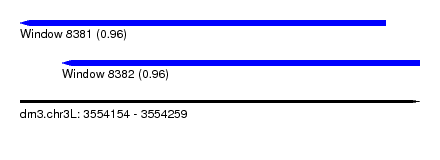
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,554,154 – 3,554,259 |
| Length | 105 |
| Max. P | 0.961034 |

| Location | 3,554,154 – 3,554,250 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 68.62 |
| Shannon entropy | 0.63241 |
| G+C content | 0.37314 |
| Mean single sequence MFE | -21.43 |
| Consensus MFE | -7.76 |
| Energy contribution | -9.34 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3554154 96 - 24543557 AAAUACUUAAGCCAAAUAUGGUUGGAUUUCCAGCCAAUCAAGACCAUUAGUAAGAUAUUCAGGGGCUUACCUGUGGCACUCAAACUAUAUUUUACA-- ..................(((((((....))))))).............(((((((((...((((((.......))).))).....))))))))).-- ( -22.90, z-score = -1.87, R) >droSim1.chr3L 3092494 96 - 22553184 AAAUACUCAGGCCAAAUAUGGUUGGAUUUCCAGCCAAUCAAGACCAUUAGUCAGAUAUUCAGCGGCUUACCUGUGGCACUCAAACUAUAUUUUACA-- ........(((((.....(((((((....))))))).....(((.....)))...........)))))...(((((........))))).......-- ( -21.40, z-score = -1.53, R) >droSec1.super_2 3545657 96 - 7591821 AAAUACUCAAGCCAAAUAUGGUUGGAUUUCCAGCCAAUCAAGACCAUUAGUCAGAUAUUCAGGGGCUUACCUGUGGCACUCAAACUUUAUUUUACA-- ..........((((....(((((((....))))))).....(((.....))).......((((......))))))))...................-- ( -24.30, z-score = -2.42, R) >droYak2.chr3L 4106277 96 - 24197627 AAAUACUCAAGCCAAAUAUGGCUGCAUUUCCAGCCAAUCAAGACCAUUAGUCAGAUGUUCAGGGGCCUACCUGUGGCAAUCAAACUAUAUUUUACA-- ..........((((....((((((......)))))).....(((.....))).......((((......))))))))...................-- ( -22.20, z-score = -1.31, R) >droEre2.scaffold_4784 6250157 96 - 25762168 AAAUACUCAAGCCAAAUACGGUUGGAUUUCCAGCCAAUCAAGACCAUUAGUCAGAUGGUCAGGGGCUUACGUGUGGCAAUCAAACUAUAUUUUACA-- ..((((..(((((......((((((....))))))......(((((((.....)))))))...)))))..))))......................-- ( -29.30, z-score = -3.83, R) >droAna3.scaffold_13337 13406624 80 + 23293914 AAAUAUUU-----AAAUAU---UGGGCUUCAA---AAACAAGACCAUUAGUCAGUCAGCCAGA-------CAGUUUAAAUCAAACUAUUUUUUAGCCU ........-----......---.(((((..((---((....(((.....))).(((.....))-------)(((((.....))))).))))..))))) ( -12.60, z-score = -1.98, R) >dp4.chrXR_group8 3524077 82 + 9212921 ------------AACCUUUGAAGACAAUUCAGAUAUGAAAAUAUCAGUUGUAAAAUAUUCAAA----CUGUCGCCCAAAUAUGGCCACACUUUUGGCG ------------....((((((.((((((..(((((....)))))))))))......))))))----....((((.(((..((....))..))))))) ( -17.30, z-score = -2.44, R) >consensus AAAUACUCAAGCCAAAUAUGGUUGGAUUUCCAGCCAAUCAAGACCAUUAGUCAGAUAUUCAGGGGCUUACCUGUGGCAAUCAAACUAUAUUUUACA__ ........(((((.....(((((((....))))))).....(((.....)))...........))))).............................. ( -7.76 = -9.34 + 1.58)
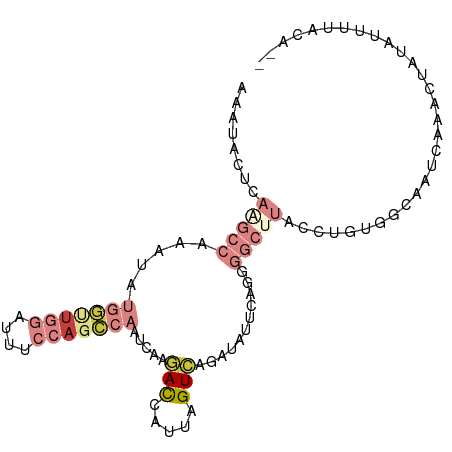
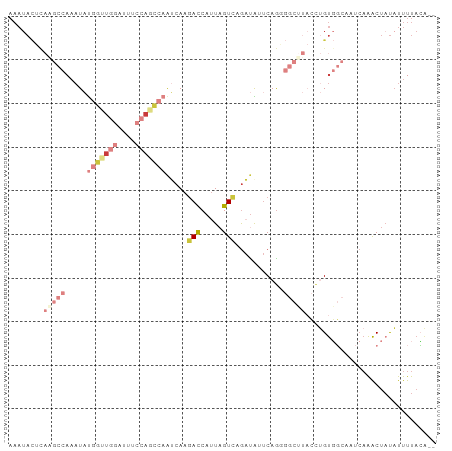
| Location | 3,554,165 – 3,554,259 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Shannon entropy | 0.47175 |
| G+C content | 0.39842 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.33 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3554165 94 - 24543557 AUGUCAAUUAAAUACUUAAGCCAAAUAUGGUUGGAUUUCCAGCCA-AUCAAGACCAUUAGUAAGAUAUUCAGGGGCUUACCUGUG-GCACUCAAAC ...................((((....(((((((....)))))))-.......................((((......))))))-))........ ( -22.80, z-score = -1.67, R) >droSim1.chr3L 3092505 94 - 22553184 AUGUCAAUUAAAUACUCAGGCCAAAUAUGGUUGGAUUUCCAGCCA-AUCAAGACCAUUAGUCAGAUAUUCAGCGGCUUACCUGUG-GCACUCAAAC .(((((...........(((((.....(((((((....)))))))-.....(((.....)))...........))))).....))-)))....... ( -21.89, z-score = -1.50, R) >droSec1.super_2 3545668 94 - 7591821 AUGUCAAUUAAAUACUCAAGCCAAAUAUGGUUGGAUUUCCAGCCA-AUCAAGACCAUUAGUCAGAUAUUCAGGGGCUUACCUGUG-GCACUCAAAC ...................((((....(((((((....)))))))-.....(((.....))).......((((......))))))-))........ ( -24.30, z-score = -2.17, R) >droYak2.chr3L 4106288 94 - 24197627 AUGUCAAUUAAAUACUCAAGCCAAAUAUGGCUGCAUUUCCAGCCA-AUCAAGACCAUUAGUCAGAUGUUCAGGGGCCUACCUGUG-GCAAUCAAAC ...................((((....((((((......))))))-.....(((.....))).......((((......))))))-))........ ( -22.20, z-score = -1.16, R) >droEre2.scaffold_4784 6250168 94 - 25762168 AUGUCAAUUAAAUACUCAAGCCAAAUACGGUUGGAUUUCCAGCCA-AUCAAGACCAUUAGUCAGAUGGUCAGGGGCUUACGUGUG-GCAAUCAAAC .(((((......(((..(((((......((((((....)))))).-.....(((((((.....)))))))...)))))..)))))-)))....... ( -30.80, z-score = -4.28, R) >dp4.chrXR_group8 3524085 83 + 9212921 -------------AUUUUGGACAACCUUUGAAGACAAUUCAGAUAUGAAAAUAUCAGUUGUAAAAUAUUCAAACUGUCGCCCAAAUAUGGCCACAC -------------......((((...((((((.((((((..(((((....)))))))))))......)))))).))))(((.......)))..... ( -17.40, z-score = -2.40, R) >consensus AUGUCAAUUAAAUACUCAAGCCAAAUAUGGUUGGAUUUCCAGCCA_AUCAAGACCAUUAGUCAGAUAUUCAGGGGCUUACCUGUG_GCACUCAAAC .................(((((......((((((....)))))).......(((.....)))...........))))).................. (-11.80 = -12.33 + 0.53)

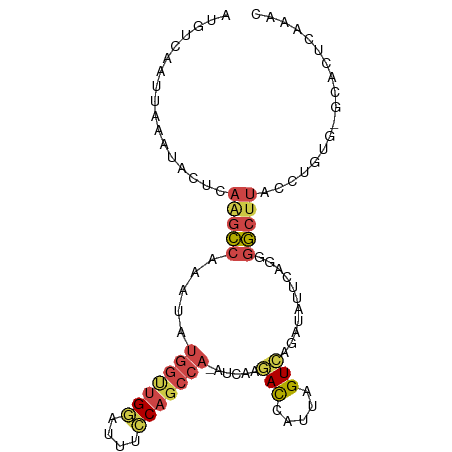
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:06 2011