| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,263,989 – 4,264,092 |
| Length | 103 |
| Max. P | 0.662446 |

| Location | 4,263,989 – 4,264,092 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.00 |
| Shannon entropy | 0.41237 |
| G+C content | 0.52770 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -17.88 |
| Energy contribution | -19.19 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
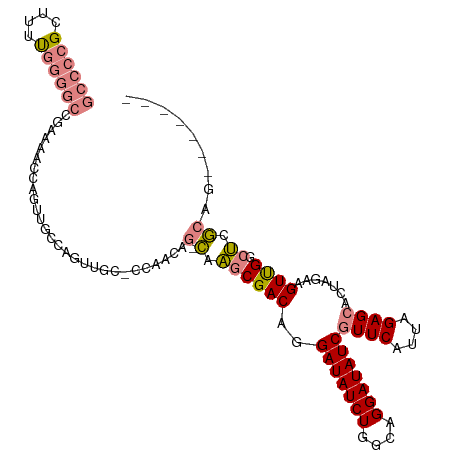
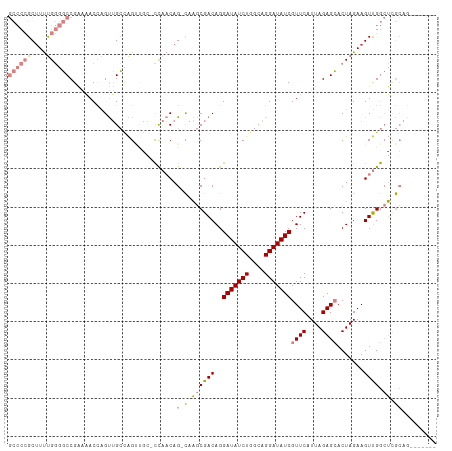
>dm3.chr2L 4263989 103 - 23011544 GCCCCGCUUUUGGGGCCGAAAACCAGUUGCCAGUUGG-CCAACAG-CAAGCGACAGGAUAUCUGGCAGGAUAUCGUUCAUUAGAGCACUAGAAGUUGGCUCGCAG------- ((((((....)))))).(((.....(((((..((((.-....)))-)..)))))..(((((((....))))))).)))....((((.(........)))))....------- ( -35.30, z-score = -1.52, R) >droPer1.super_8 332530 107 + 3966273 -----GCUUUCGCCUCCGUCUACUUGCGGAGGGACCCGUUACCAGUUACCCGACAGGAUAUCUGACAGGAUAUCGUUCAUUAGAGAACUAGAAGUCGAAGUAGAAGUAGCAG -----....((.(((((((......)))))))))...(((((...((((.((((..(((((((....)))))))((((......)))).....))))..))))..))))).. ( -35.90, z-score = -2.85, R) >dp4.chr4_group2 255261 107 - 1235136 -----GCCUCCGCCUCCGUCUACUUGCGGAUGGACCCGUUACCAGUUACCCGACAGGAUAUCUGACAGGAUAUCGUUCAUUAGAGAACUAGAAGUCGAAGUAGAAGUAGCAG -----...((((..(((((......)))))))))...(((((...((((.((((..(((((((....)))))))((((......)))).....))))..))))..))))).. ( -31.80, z-score = -1.93, R) >droSim1.chr2L 4184510 103 - 22036055 GCCCCGCUUUUGGGGCCGAAAACCAGUUGCCAGUUGG-CCAACAG-CAAGCGACAGGAUAUCUGGCAGGAUAUCGUUCAUUAGAGCACUAGAAGUUGGCUCGCAG------- ((((((....)))))).(((.....(((((..((((.-....)))-)..)))))..(((((((....))))))).)))....((((.(........)))))....------- ( -35.30, z-score = -1.52, R) >droSec1.super_5 2362085 103 - 5866729 GCCCCGCUUUUGGGGCCGAAAACCAGUUGCCAGUUGG-CCAACAG-CAAGCGACAGGAUAUCUGGCAGGAUAUCGUUCAUUAGAGCACUAGAAGUUGGCUCGCAG------- ((((((....)))))).(((.....(((((..((((.-....)))-)..)))))..(((((((....))))))).)))....((((.(........)))))....------- ( -35.30, z-score = -1.52, R) >droYak2.chr2L 4292014 104 - 22324452 GCCCCGCUUUUGGGGCCGAAAACCAGUUGCCAGCAGAUCCAACAG-CAAGCGACAGGAUAUCUGGCAGGAUAUCGUUCAUUAGAGCACUAGAAGUUGGCUCGCAA------- ((((((....))))))(((...((...((((((...((((....(-....)....))))..))))))))...))).......((((.(........)))))....------- ( -31.90, z-score = -0.93, R) >droEre2.scaffold_4929 4340378 103 - 26641161 GCCCCGCUUUUGGGGCCGAAAACCAGUUGCCAGUUAC-CCAACAG-CAAGCGACAGGAUAUCUGGCAGGAUAUCGUUCAUUAGAGCACUAGAAGUUGGCUGGCAG------- ((((((....))))))...........((((((....-(((((..-..........(((((((....)))))))((((....)))).......))))))))))).------- ( -36.50, z-score = -2.32, R) >consensus GCCCCGCUUUUGGGGCCGAAAACCAGUUGCCAGUUGC_CCAACAG_CAAGCGACAGGAUAUCUGGCAGGAUAUCGUUCAUUAGAGCACUAGAAGUUGGCUCGCAG_______ ((((((....))))))..................................((((..(((((((....)))))))((((....)))).......))))............... (-17.88 = -19.19 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:53 2011