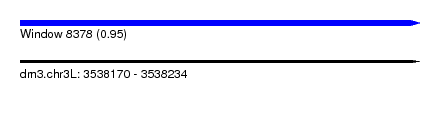
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,538,170 – 3,538,234 |
| Length | 64 |
| Max. P | 0.950420 |

| Location | 3,538,170 – 3,538,234 |
|---|---|
| Length | 64 |
| Sequences | 5 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 90.58 |
| Shannon entropy | 0.16107 |
| G+C content | 0.42565 |
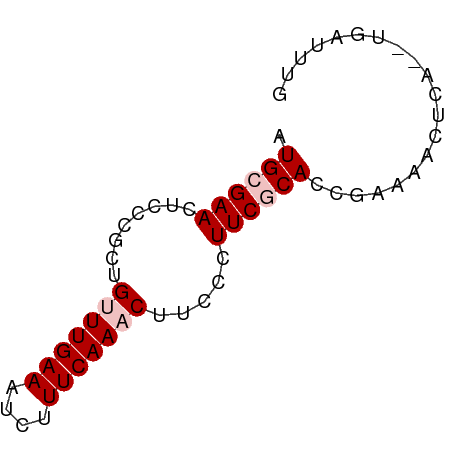
| Mean single sequence MFE | -9.17 |
| Consensus MFE | -6.96 |
| Energy contribution | -7.76 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
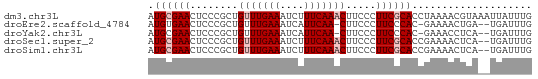
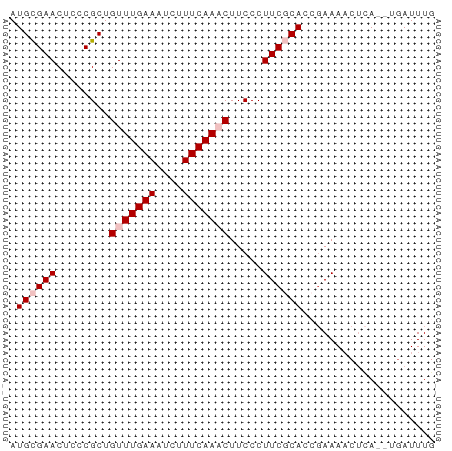
>dm3.chr3L 3538170 64 + 24543557 AUGCGAACUCCCGCUGUUUGAAAUCUUUCAAACUUCCCUUCGCACCUAAAACGUAAAUUAUUUG .((((((........(((((((....))))))).....)))))).................... ( -11.32, z-score = -4.51, R) >droEre2.scaffold_4784 6232645 60 + 25762168 AUGUGAACUCCCGCUGUUUGAAAUCAUUCAA-CUUCCCUUCCCAC-GAAAACUGA--UGAUUUG ..(((......)))......((((((((...-.(((.........-)))....))--)))))). ( -5.70, z-score = -0.70, R) >droYak2.chr3L 4089930 60 + 24197627 AUGCGAACUCCCGCUGUUUGAAAUCAUUCAA-CUUCCCUUCCCAC-GAAACCUCA--UGAUUUG ...(((((.......)))))(((((((....-.(((.........-))).....)--)))))). ( -6.20, z-score = -1.39, R) >droSec1.super_2 3529879 62 + 7591821 AUGCGAACUCCCGCUGUUUGAAAUCUUUCAAACUUCCCUUCGCACCGAAAACUCA--UGAUUUG .((((((........(((((((....))))))).....))))))...........--....... ( -11.32, z-score = -3.49, R) >droSim1.chr3L 3076283 62 + 22553184 AUGCGAACUCCCGCUGUUUGAAAUCUUUCAAACUUCCCUUCGCACCGAAAACUCA--UGAUUUG .((((((........(((((((....))))))).....))))))...........--....... ( -11.32, z-score = -3.49, R) >consensus AUGCGAACUCCCGCUGUUUGAAAUCUUUCAAACUUCCCUUCGCACCGAAAACUCA__UGAUUUG .((((((........(((((((....))))))).....)))))).................... ( -6.96 = -7.76 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:01:03 2011