| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,474,987 – 3,475,080 |
| Length | 93 |
| Max. P | 0.830871 |

| Location | 3,474,987 – 3,475,080 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.99 |
| Shannon entropy | 0.56670 |
| G+C content | 0.51036 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -9.64 |
| Energy contribution | -10.64 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
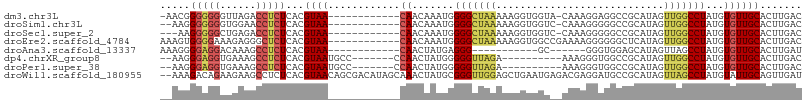
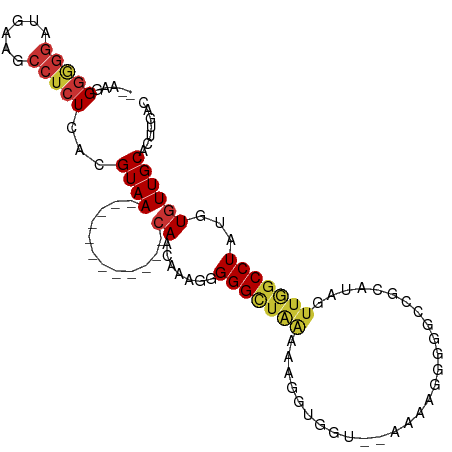
>dm3.chr3L 3474987 93 - 24543557 -AACGGGGGGGUUAGACCUCUCACGUAA------------CAACAAAUGGGGCUAAAAAGGUGGUA-CAAAGGAGGCCGCAUAGUUGGCCUAUGUGUUGCACUUGAC -.(((((((((.....)))))).)))..------------(((((....(((((((....(((((.-........)))))....)))))))...)))))........ ( -33.60, z-score = -3.06, R) >droSim1.chr3L 3013006 92 - 22553184 --AAGGGGGGGUGGAACCUCUCACGUAA------------CAACAAAUGGGGCUAAAAAGGUGGUC-CAAAGGGGGCCGCAUAGUUGGCCUAUGUGUUGCACUUGAC --..((((((......))))))..((((------------((.((....(((((((....((((((-(.....)))))))....))))))).))))))))....... ( -39.30, z-score = -4.19, R) >droSec1.super_2 3467658 91 - 7591821 ---AAGGGGGCUGAGACCUCUCACGUAA------------CAACAAAUGGGGCUAAAAAGGUGGUC-CAAAGGGGGCCGCAUAGUUGGCCUAUGUGUUGCACUUGAC ---.(((((.......)))))...((((------------((.((....(((((((....((((((-(.....)))))))....))))))).))))))))....... ( -35.30, z-score = -2.82, R) >droEre2.scaffold_4784 6167372 95 - 25762168 AAAGUGGGGAAAGAGGGCUCUCACGUAA------------CAACAAAUGGGGCUAAAAAGGUGGCCGAAAAGGGGGGCUCAUAGUUGGCCUAUGUGUUGCACUUGAC .(((((((((........))))......------------(((((....(((((((....((((((.........))).)))..)))))))...))))))))))... ( -29.10, z-score = -2.06, R) >droAna3.scaffold_13337 13328681 78 + 23293914 AAAGGGGAGGACAAAGCCUCUCACGUAA------------CAACUAUGAGGG-----------GC------GGGUGGAGCAUAGUUAGCCUAUGUGUUGCACUUGAU ...............((((((((.((..------------..))..))))))-----------))------(((((.((((((.........)))))).)))))... ( -23.30, z-score = -1.31, R) >dp4.chrXR_group8 5587598 88 - 9212921 --AAGGGAGGUGAAAGCCUCUCACGUAAUGCC-------CCAACUAUGGGGGUUAGA----------AAAGGGUGGCCGCAUAGUUGGCCUAUGUGUUGCACUUGAC --..(((((((....)))))))..(((((((.-------(((((((((.((.(....----------.......).)).))))))))).....)))))))....... ( -34.30, z-score = -2.48, R) >droPer1.super_38 703763 88 + 801819 --AAGGGAGGUGAAAGCCUCUCACGUAAUGCC-------CCAACUAUGGGGGUUAGA----------AAAGGGUGGCCGCAUAGUUGGCCUAUGUGUUGCACUUGAC --..(((((((....)))))))..(((((((.-------(((((((((.((.(....----------.......).)).))))))))).....)))))))....... ( -34.30, z-score = -2.48, R) >droWil1.scaffold_180955 1232811 105 - 2875958 --AAAGACAGAAGAAGCCUCUCACGUAACAGCGACAUAGCAAACUAUGCGGGUUGGAGCUGAAUGAGACGAGGAUGCCGCAUAGUUAGCCUAUGUAUUGCAGUUGAU --...(((........((((((.(((..((((..((..(((.....)))....))..)))).))).)).)))).(((.((((((.....))))))...))))))... ( -27.20, z-score = -0.85, R) >consensus __AAGGGGGGAUGAAGCCUCUCACGUAA____________CAACAAAGGGGGCUAAAAAGGUGGU__AAAAGGGGGCCGCAUAGUUGGCCUAUGUGUUGCACUUGAC .....(((((......)))))...((((......................((((....................))))((((((.....)))))).))))....... ( -9.64 = -10.64 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:53 2011