| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,463,912 – 3,464,009 |
| Length | 97 |
| Max. P | 0.936471 |

| Location | 3,463,912 – 3,464,009 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 64.82 |
| Shannon entropy | 0.64632 |
| G+C content | 0.41317 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
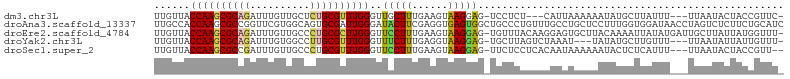
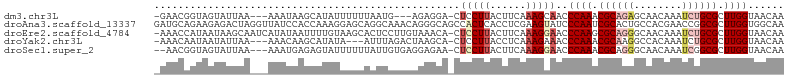
>dm3.chr3L 3463912 97 + 24543557 UUGUUACCAAGCGCAGAUUUGUUGCUCUGCGUUUGGGUUGCUUUGAAGUAAGGAG-UCCUCU---CAUUAAAAAAUAUGCUUAUUU---UUAAUACUACCGUUC- ..((..(((((((((((........)))))))))))...)).....((((..(((-....))---)....(((((((....)))))---))..)))).......- ( -25.20, z-score = -2.52, R) >droAna3.scaffold_13337 13317834 105 - 23293914 UUGCCACCAAGCGCCGGUUCGUGGCAGUGCGAUUGGGAUACUUCGAGGUGAGUGGCUGCCCUGUUUGCCUGCUCCUUUGGUGGAUAACCUAGUCUCUUCUGCAUC .((((((..(((....))).))))))(((((((((((.((((((((((.(((((((..........))).)))))))))).)).)).)))))))......)))). ( -33.50, z-score = 0.09, R) >droEre2.scaffold_4784 6156059 103 + 25762168 UUGUUACCAAGCGCAGAUUUGUUGCCCUGCGCUUGGGUUCCUUUGAAGUAAGGAG-UGUUUACAAGGAGUGCUUACAAAAUUAUAUGAUUGCUUAUUAUGGUUU- ......((((((((((..........))))))))))..(((((......)))))(-((..(((.....)))..))).((((((((...........))))))))- ( -27.40, z-score = -1.32, R) >droYak2.chr3L 4013039 97 + 24197627 UUGUUACCAAGCGCAGAUUUGUGGCCUUGCGUUUGGGUUUCUUUGAGGUAAGGAG-UGCUUAGUCUAAAU---UAUAUGCUUGUUU---UUAAUAUUAUUGUUU- ......((((((((((..........))))))))))......(((((..((.(((-((..((((....))---))..))))).)))---))))...........- ( -17.40, z-score = 0.65, R) >droSec1.super_2 3456505 99 + 7591821 UUGUUACCAAGCGCCGAUUUGUUGCCCUGCGUUUGGGUUCCUUUGAAGUAAGGAG-UUCUCCUCACAAUAAAAAAUACUCUCAUUU---UUAAUACUACCGUU-- (((((.((((((((.(..........).))))))))...........((.((((.-...)))).)))))))...............---..............-- ( -17.80, z-score = -0.41, R) >consensus UUGUUACCAAGCGCAGAUUUGUUGCCCUGCGUUUGGGUUCCUUUGAAGUAAGGAG_UGCUCAGUACAAUUAAAAAUAUGCUUAUUU___UUAAUACUACCGUUU_ ......((((((((((..........))))))))))..(((((......)))))................................................... (-15.90 = -16.02 + 0.12)
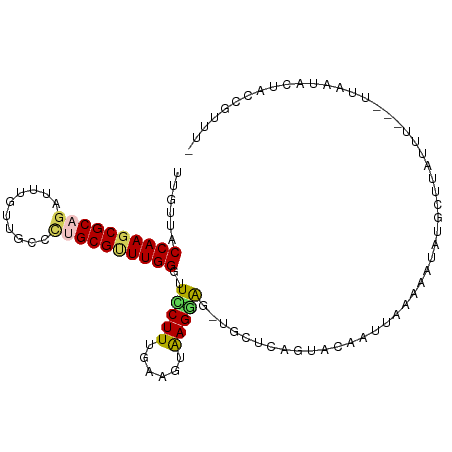
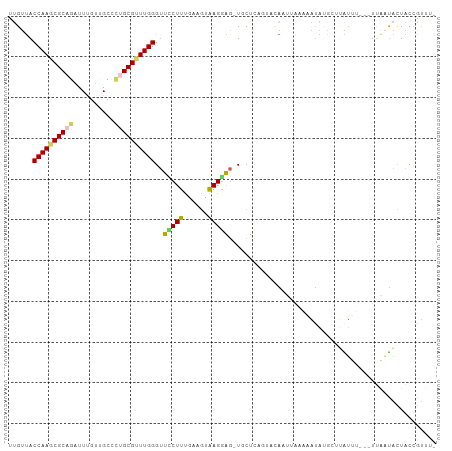
| Location | 3,463,912 – 3,464,009 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 64.82 |
| Shannon entropy | 0.64632 |
| G+C content | 0.41317 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -9.48 |
| Energy contribution | -11.00 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
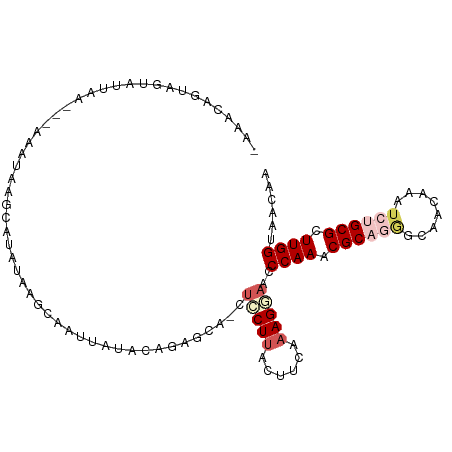
>dm3.chr3L 3463912 97 - 24543557 -GAACGGUAGUAUUAA---AAAUAAGCAUAUUUUUUAAUG---AGAGGA-CUCCUUACUUCAAAGCAACCCAAACGCAGAGCAACAAAUCUGCGCUUGGUAACAA -....(((.(((((((---(((.........)))))))).---.((((.-.......))))...)).)))(((.((((((........)))))).)))....... ( -19.80, z-score = -1.77, R) >droAna3.scaffold_13337 13317834 105 + 23293914 GAUGCAGAAGAGACUAGGUUAUCCACCAAAGGAGCAGGCAAACAGGGCAGCCACUCACCUCGAAGUAUCCCAAUCGCACUGCCACGAACCGGCGCUUGGUGGCAA (((((....(((....(((.....)))....(((..(((..........))).)))..)))...)))))..........((((((.((.......)).)))))). ( -24.90, z-score = 1.29, R) >droEre2.scaffold_4784 6156059 103 - 25762168 -AAACCAUAAUAAGCAAUCAUAUAAUUUUGUAAGCACUCCUUGUAAACA-CUCCUUACUUCAAAGGAACCCAAGCGCAGGGCAACAAAUCUGCGCUUGGUAACAA -............((...((........))...))......(((.....-.(((((......)))))..(((((((((((........)))))))))))..))). ( -26.00, z-score = -3.68, R) >droYak2.chr3L 4013039 97 - 24197627 -AAACAAUAAUAUUAA---AAACAAGCAUAUA---AUUUAGACUAAGCA-CUCCUUACCUCAAAGAAACCCAAACGCAAGGCCACAAAUCUGCGCUUGGUAACAA -...............---.............---......(((((((.-...(((......)))..........(((.((.......))))))))))))..... ( -10.70, z-score = -0.49, R) >droSec1.super_2 3456505 99 - 7591821 --AACGGUAGUAUUAA---AAAUGAGAGUAUUUUUUAUUGUGAGGAGAA-CUCCUUACUUCAAAGGAACCCAAACGCAGGGCAACAAAUCGGCGCUUGGUAACAA --......((((..((---(.((....)).)))..))))(((((((...-.)))))))...........((((.(((..(....)......))).))))...... ( -20.30, z-score = -0.32, R) >consensus _AAACAGUAGUAUUAA___AAAUAAGCAUAUAAGCAAUUAUACAGAGCA_CUCCUUACUUCAAAGGAACCCAAACGCAGGGCAACAAAUCUGCGCUUGGUAACAA ...................................................(((((......)))))..((((.((((((........)))))).))))...... ( -9.48 = -11.00 + 1.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:49 2011