| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,452,746 – 3,452,854 |
| Length | 108 |
| Max. P | 0.946029 |

| Location | 3,452,746 – 3,452,854 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.42 |
| Shannon entropy | 0.61300 |
| G+C content | 0.38866 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -8.60 |
| Energy contribution | -9.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
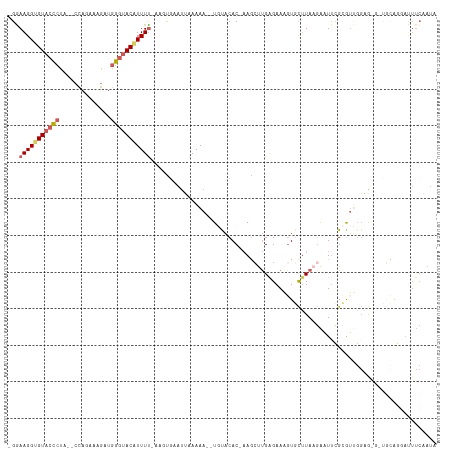
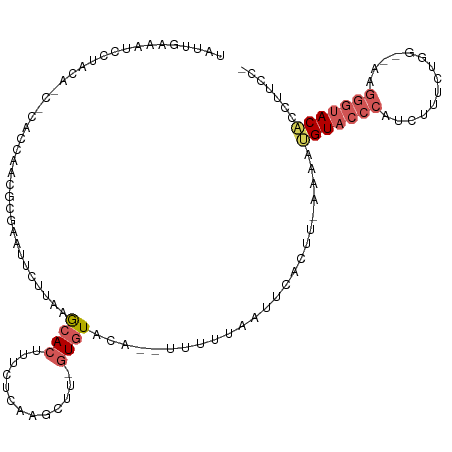
>dm3.chr3L 3452746 108 + 24543557 -GGAAGGUGUUGUAC----CAAAAAGAUGGGUACAUUUC--UUUGAAUUGAAAAUAUCUACAC-AAGCUUGAGAAAGUGCUUAAGAAUAAGCGUUGGAGUG-UGUAUGAUUUCAAUA -.(((((...(((((----(.........))))))..))--)))..(((((((.(((.(((((-..((((....))))(((((....)))))......)))-)).))).))))))). ( -28.40, z-score = -3.13, R) >droSim1.chr3L 2991141 107 + 22553184 -GGAAGGUGUACCCUA--CCAGAAAGAUGGGUACAUUUU-ACGCGAAUUAAAAA--UGUACAC-AAGCUUGAGAAAGUGCUUAAGAAUUCGCGUUGGAG---UGCAGGAUUUCGAUA -..(((((((((((..--..........)))))))))))-..(((((((.....--((....)-)..((((((......)))))))))))))(((((((---(.....)))))))). ( -30.90, z-score = -3.04, R) >droSec1.super_2 3445323 114 + 7591821 GGAAAGGUGUACCCCUACCCAGAAUGAUGGGUACAUUUUUAAGCGAAUUAAAA---UCUACACGAAGCUUGAGAACGUGCUUAUGAUUUCGGAGGGGGGAGGUGCAGGAUUUCAAUA .((((..(((((((((.(((..(((.((((((((.(((((((((.........---..........))))))))).)))))))).)))..)).)..))).))))))...)))).... ( -39.51, z-score = -3.99, R) >droYak2.chr3L 4001328 109 + 24197627 -GGAAGGAGUACCCUA--CUAGAAAGAUGGGUACUUUUU-AAAUUCAUAAAAAA--UGCACAC--AAGUCGGGAAAGUGCUUAAGAAUUCGCGUUGGUAGUGCUUAGAACUUCUAUG -(((((((((((((..--((....))..))))))))...-..............--.((((((--....((.(((..(......)..))).))...)).))))......)))))... ( -26.00, z-score = -1.35, R) >droEre2.scaffold_4784 6144581 93 + 25762168 -GGAAGGUGUACCCUU--CUAUAAAGAUGGGUACUUUUU-AAGUGAAUUUAAAA--UGCACAC-AAGCCUGAGAAA-UUUGUAUCGCUUGGGAUCUUUACA---------------- -.((((((((((((.(--((....))).))))))..(((-((((((((((....--.((....-..)).....)))-).....)))))))))))))))...---------------- ( -22.70, z-score = -1.61, R) >consensus _GGAAGGUGUACCCUA__CCAGAAAGAUGGGUACAUUUU_AAGUGAAUUAAAAA__UGUACAC_AAGCUUGAGAAAGUGCUUAAGAAUUCGCGUUGGAG_G_UGCAGGAUUUCAAUA ...(((((((((((..............))))))))))).............................................................................. ( -8.60 = -9.20 + 0.60)


| Location | 3,452,746 – 3,452,854 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.42 |
| Shannon entropy | 0.61300 |
| G+C content | 0.38866 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -5.58 |
| Energy contribution | -6.34 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.838913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
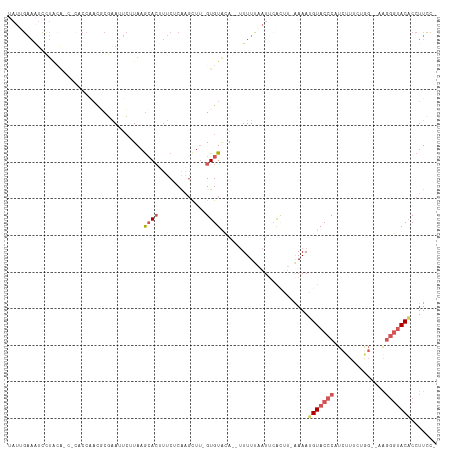
>dm3.chr3L 3452746 108 - 24543557 UAUUGAAAUCAUACA-CACUCCAACGCUUAUUCUUAAGCACUUUCUCAAGCUU-GUGUAGAUAUUUUCAAUUCAAA--GAAAUGUACCCAUCUUUUUG----GUACAACACCUUCC- .(((((((...((((-((.......(((((....))))).(((....)))..)-))))).....))))))).....--....((((((.........)----))))).........- ( -20.40, z-score = -3.70, R) >droSim1.chr3L 2991141 107 - 22553184 UAUCGAAAUCCUGCA---CUCCAACGCGAAUUCUUAAGCACUUUCUCAAGCUU-GUGUACA--UUUUUAAUUCGCGU-AAAAUGUACCCAUCUUUCUGG--UAGGGUACACCUUCC- ....(((........---.....(((((((((.....((((............-))))...--.....)))))))))-....((((((((((.....))--).)))))))..))).- ( -25.12, z-score = -3.51, R) >droSec1.super_2 3445323 114 - 7591821 UAUUGAAAUCCUGCACCUCCCCCCUCCGAAAUCAUAAGCACGUUCUCAAGCUUCGUGUAGA---UUUUAAUUCGCUUAAAAAUGUACCCAUCAUUCUGGGUAGGGGUACACCUUUCC ....((((...((.(((((........((((((....(((((...........))))).))---))))................((((((......))))))))))).))..)))). ( -22.60, z-score = -1.08, R) >droYak2.chr3L 4001328 109 - 24197627 CAUAGAAGUUCUAAGCACUACCAACGCGAAUUCUUAAGCACUUUCCCGACUU--GUGUGCA--UUUUUUAUGAAUUU-AAAAAGUACCCAUCUUUCUAG--UAGGGUACUCCUUCC- ((((((((......((((......((.(((............))).))....--..)))).--.)))))))).....-....(((((((..((....))--..)))))))......- ( -20.10, z-score = -1.27, R) >droEre2.scaffold_4784 6144581 93 - 25762168 ----------------UGUAAAGAUCCCAAGCGAUACAAA-UUUCUCAGGCUU-GUGUGCA--UUUUAAAUUCACUU-AAAAAGUACCCAUCUUUAUAG--AAGGGUACACCUUCC- ----------------(((((((((..(((((........-........))))-).((((.--((((((......))-)))).))))..)))))))))(--((((.....))))).- ( -22.59, z-score = -3.19, R) >consensus UAUUGAAAUCCUACA_C_CACCAACGCGAAUUCUUAAGCACUUUCUCAAGCUU_GUGUACA__UUUUUAAUUCACUU_AAAAUGUACCCAUCUUUCUGG__AAGGGUACACCUUCC_ ..................................................................................(((((((..............)))))))....... ( -5.58 = -6.34 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:48 2011