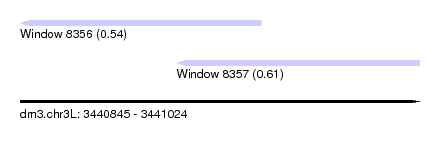
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,440,845 – 3,441,024 |
| Length | 179 |
| Max. P | 0.612814 |

| Location | 3,440,845 – 3,440,953 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.73 |
| Shannon entropy | 0.12053 |
| G+C content | 0.50111 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
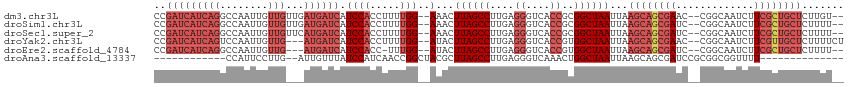
>dm3.chr3L 3440845 108 - 24543557 UAAUUAAGCAGCGAACCGGCAAUCUUCGCUGCUC------------UUGUAUACUUUUUAAGUCCGCGGCCGUUGUUUAUCCUCCGGUCUCAUCAGAUCCGCCUCGAUCAGCCCCCGUGC ......(((((((((.........))))))))).------------..................(((((..((((...(((...((((((....))).)))....)))))))..))))). ( -26.90, z-score = -1.37, R) >droSim1.chr3L 2978409 108 - 22553184 UAAUUAAGCAGCGAUCCGGCAAUCUUCGCUGCUC------------UUUUAUACUUUUUAAGUCCGCGGCCGUUGUUUAUCCUCCGGUCUCAUCAGAUCCGCCUCGAUCAGCCCCCGUGC ......((((((((...........)))))))).------------..................(((((..((((...(((...((((((....))).)))....)))))))..))))). ( -26.70, z-score = -1.63, R) >droSec1.super_2 3432943 108 - 7591821 UAAUUAAGCAGCGAUCCGGCAAUCUUCGCUGCUC------------UUUUAUACUUUUUAAGUCCGCGGCCGUUGUUUAUCCUCCGGUCUCAUCAGAUCCGCCUUGAUCAGCCCCCGUGC ......((((((((...........)))))))).------------..................(((((..((((.(((.....((((((....))).)))...))).))))..))))). ( -27.60, z-score = -1.97, R) >droYak2.chr3L 3988790 120 - 24197627 UAAUUAAGCAGCGAACCGGCAAUCUUCGUUGCUCUUUUCUUUUUUUUUUUAUACAUUUUAAGUCUGCAGCCGUUGUUUAUCCUCCGGUCUCAUCAGAUCCGCCUCGAUCAGCCCCCGUGC ......(((((((((.........)))))))))..................(((..........((..((((..(......)..))))..))...((((......)))).......))). ( -18.80, z-score = 0.10, R) >droEre2.scaffold_4784 6130820 108 - 25762168 UAAUUAAGCAGCGAUCCGGCAAUCUUCGCUGCUC------------UUUUAUACUUUUUAAGUCUGCAGCCGUUGUUUAUCCUCCGGUCUCAUCAGAUCCGCCUCGAUCAGCCCCCGUGC .......(((((((((.(((.......(((((..------------.((((.......))))...)))))...............(((((....))))).)))..)))).)).....))) ( -23.10, z-score = -0.95, R) >consensus UAAUUAAGCAGCGAUCCGGCAAUCUUCGCUGCUC____________UUUUAUACUUUUUAAGUCCGCGGCCGUUGUUUAUCCUCCGGUCUCAUCAGAUCCGCCUCGAUCAGCCCCCGUGC ......((((((((...........))))))))...............................(((((..((((...(((...((((((....))).)))....)))))))..))))). (-22.70 = -22.70 + -0.00)


| Location | 3,440,915 – 3,441,024 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Shannon entropy | 0.35568 |
| G+C content | 0.48514 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -16.99 |
| Energy contribution | -19.38 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3440915 109 - 24543557 CCGAUCAUCAGGCCAAUUGUUGUUGAUGAUCAUCCACCUUUUGG--AAACUUAGCCUUGAGGGUCACCGCGGCUAAUUAAGCAGCGAAC--CGGCAAUCUUCGCUGCUCUUGU-- ..(((((((((((.....))..))))))))).((((.....)))--)...((((((....((....))..))))))...(((((((((.--........))))))))).....-- ( -35.00, z-score = -2.01, R) >droSim1.chr3L 2978479 109 - 22553184 CCGAUCAUCAGGCCAAUUGUUGUUGAUGAUCAUCCACCUUUUGG--AAACUUAGCCUUGAGGGUCACCGCGGCUAAUUAAGCAGCGAUC--CGGCAAUCUUCGCUGCUCUUUU-- ..(((((((((((.....))..))))))))).((((.....)))--)...((((((....((....))..))))))...((((((((..--.........)))))))).....-- ( -34.80, z-score = -2.18, R) >droSec1.super_2 3433013 109 - 7591821 CCGAUCAUCAGGCCAAUUGUUGUUCAUGAUCAUCCACCUUUUGG--AAACUUAGCCUUGAGGGUCACCGCGGCUAAUUAAGCAGCGAUC--CGGCAAUCUUCGCUGCUCUUUU-- ..((((((..(..((.....))..))))))).((((.....)))--)...((((((....((....))..))))))...((((((((..--.........)))))))).....-- ( -30.80, z-score = -1.31, R) >droYak2.chr3L 3988870 108 - 24197627 CCGAUCAUCAGUCCAAUUGUUG---AUGAUCAUCCACCUUUUGG--AUACUUAGCCUUGAGGGUCACCGUGGCUAAUUAAGCAGCGAAC--CGGCAAUCUUCGUUGCUCUUUUCU ..(((((((((........)))---))))))(((((.....)))--))..((((((....((....))..))))))...(((((((((.--........)))))))))....... ( -35.20, z-score = -3.51, R) >droEre2.scaffold_4784 6130890 105 - 25762168 CCGAUCAUCAGGCCAAUUGUUG---AUGAUCAUCCACC-UUUGG--AUACUUAGCCUUGAGGGUCACCGUGGCUAAUUAAGCAGCGAUC--CGGCAAUCUUCGCUGCUCUUUU-- ..(((((((((........)))---))))))(((((..-..)))--))..((((((....((....))..))))))...((((((((..--.........)))))))).....-- ( -37.30, z-score = -3.39, R) >droAna3.scaffold_13337 13294875 87 + 23293914 ------------CCAUUCCUUG--AUUGUUUAUCCAUCAACCGGCUACGCUUAGCCUUGAGGGUCAAACUGGCUAAUUAAGCAGCGAUCCGCGGCGGUUUU-------------- ------------((........--...((((((((.((((..(((((....))))))))))))).))))...........((.(((...))).))))....-------------- ( -20.30, z-score = 0.49, R) >consensus CCGAUCAUCAGGCCAAUUGUUG___AUGAUCAUCCACCUUUUGG__AAACUUAGCCUUGAGGGUCACCGCGGCUAAUUAAGCAGCGAUC__CGGCAAUCUUCGCUGCUCUUUU__ ..((((((.................))))))...................((((((....((....))..))))))...((((((((.............))))))))....... (-16.99 = -19.38 + 2.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:45 2011