
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,404,373 – 3,404,470 |
| Length | 97 |
| Max. P | 0.938912 |

| Location | 3,404,373 – 3,404,470 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 68.41 |
| Shannon entropy | 0.61272 |
| G+C content | 0.58680 |
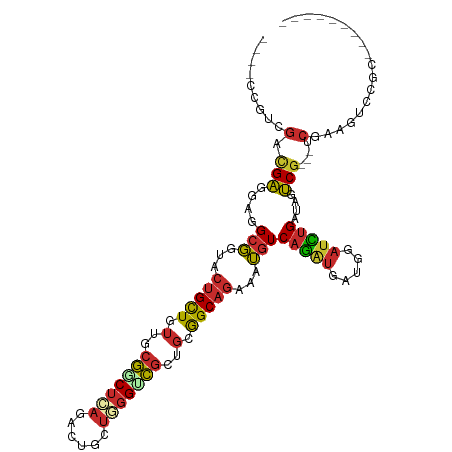
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.48 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
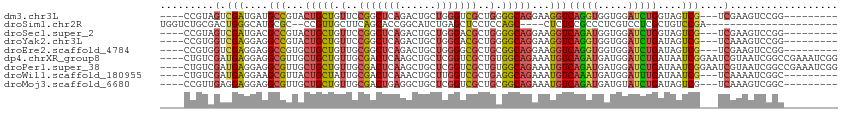
>dm3.chr3L 3404373 97 + 24543557 ----CCGUAGUCGAUGAUGCCGUACUGCUGUUCCGGCUCAGACUGCUGGGUCGCUGGGGCAGGAAGGUCAGGUGGUGGAUCUGGUAGUCG---UCGAAGUCCGG--------- ----(((...(((((((((((...(((((.(..((((((((....))))))))..).)))))...)))(((((.....)))))...))))---))))....)))--------- ( -41.50, z-score = -2.04, R) >droSim1.chr2R 10785370 85 - 19596830 UGGUCUGCGACUGGGCAUGCGC--CCGUUGCUUCAGCACCGGCAUCUGAGCUCCUCCAGC----CUCUCCGCCCUCGUCCUCGCUGUCCGA---------------------- .((.(.((((..((((....((--(.((.((....)))).)))....(((((.....)).----)))...))))......)))).).))..---------------------- ( -26.80, z-score = 0.07, R) >droSec1.super_2 3397178 97 + 7591821 ----CCGUAGUCGAUGACGCCGUACUGCUGUUCCGGCUCAGACUGCUGGGACGCUGGGGCAGGAAGGUCAGAUGGUGGAUCUGGUAGUCG---UCGAAGUCCGG--------- ----(((...(((((((((((...(((((((((((((.......)))))))).....)))))...)))(((((.....)))))...))))---))))....)))--------- ( -41.90, z-score = -1.84, R) >droYak2.chr3L 3951900 97 + 24197627 ----CCGUGGUCGAGGAGGCCGUACUGCUGUUCCGGCUCAGACUGCUGGGACGCUGGGGCAGGAAGGUCAGGUGGUGGAUCUGAUAGUCG---UCAAAGUCCGG--------- ----(((.((((.....))))..(((.((((((((((((((....))))...))))))))))((..(((((((.....)))))))..)).---....))).)))--------- ( -35.30, z-score = 0.16, R) >droEre2.scaffold_4784 6094678 97 + 25762168 ----CCGUGGUCGAGGAGGCCGUGCUGCUGUUGCGGCUCAGACUGCUGGGGCGCUGCGGCAGGAAGGUCAGGUGGUGGAUCUGAUAGUCG---UCGAAGUCCGG--------- ----(((...((((.((((((.(.(((((((.(((.(((((....))))).))).))))))).).))))((((.....)))).....)).---))))....)))--------- ( -43.50, z-score = -1.69, R) >dp4.chrXR_group8 5499339 109 + 9212921 ----CUGUCGAUGAGGAGGCGUUGCUGCUGUUGCGACUCAAGCUGCUCGGUCGCUGUGGCAGAAAUGUCAGAUGAUGGAUCUGAUAAUCGGAAUCGUAAUCGGCCGAAAUCGG ----....((((..((.((((((.((((..(.((((((..........)))))).)..)))).)))))).(((.((((.((((.....)))).)))).)))..))...)))). ( -39.40, z-score = -1.86, R) >droPer1.super_38 613707 109 - 801819 ----CUGUCGAUGAGGAGGCGUUGCUGCUGUUGCGACUCAAGCUGCUCGGUCGCUGUGGCAGAAAUGUCAGAUGAUGGAUCUGAUAAUCGGAAUCGUAAUCGGCCGAAAUCGG ----....((((..((.((((((.((((..(.((((((..........)))))).)..)))).)))))).(((.((((.((((.....)))).)))).)))..))...)))). ( -39.40, z-score = -1.86, R) >droWil1.scaffold_180955 1125964 97 + 2875958 ----CUGUCGAUGAGGAAGCGUUACUGCUAUUGCGACUCAAACUGCUUGGUCGCUGAGGCAGAAAUGUCAAAUGAUGGAUUUGAUAAUCG---UCAAAAUCGGC--------- ----..((((((((.((.......(((((...(((((.(((.....))))))))...)))))...((((((((.....)))))))).)).---)))...)))))--------- ( -31.10, z-score = -2.78, R) >droMoj3.scaffold_6680 19449108 97 - 24764193 ----CCGUUGAGGAGGAGGCGUUGCUGCUGUUGCGACUGAGGCUGCUCGGUCGCUGCGGCAGAAAUGUCAGAUGAUGUAUCUGAUAGUCG---UCAAAGUCGGC--------- ----..(((((......((((((.(((((((.(((((((((....))))))))).))))))).)))))).((((((..........))))---))....)))))--------- ( -43.40, z-score = -3.69, R) >consensus ____CCGUCGACGAGGAGGCGGUACUGCUGUUGCGGCUCAGACUGCUGGGUCGCUGCGGCAGAAAUGUCAGAUGAUGGAUCUGAUAGUCG___UCGAAGUCCGC_________ ...........(((....(((...(((((.(..(((((((......)))))))..).)))))...)))(((((.....)))))....)))....................... (-16.61 = -16.48 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:37 2011