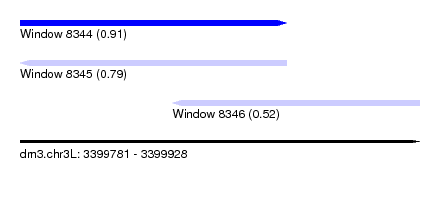
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,399,781 – 3,399,928 |
| Length | 147 |
| Max. P | 0.907489 |

| Location | 3,399,781 – 3,399,879 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.08 |
| Shannon entropy | 0.44661 |
| G+C content | 0.56896 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -16.41 |
| Energy contribution | -17.47 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
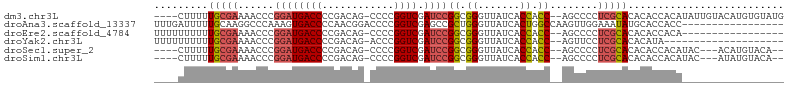
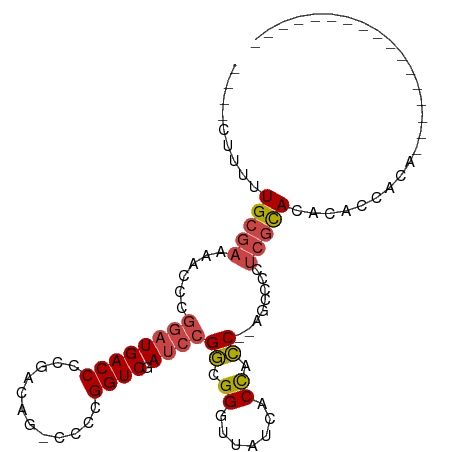
>dm3.chr3L 3399781 98 + 24543557 ----CUUUUUGCGAAAACCCGGAUGACCCCGACAG-CCCCGGUCGAUCCGGCGGGUUAUCACCACC--AGCCCCUCGCACACACCACAUAUUGUACAUGUGUAUG ----......((((....((((((((((.......-....)))).)))))).(((((.........--))))).))))(((((..((.....))...)))))... ( -27.90, z-score = -1.55, R) >droAna3.scaffold_13337 13249409 88 - 23293914 UUUGAUUUUUGCAAGGCCCAAAGUGACCCCAACGGACCCCGGUCGAGCCGCUGGGUUAUCACUGGCCAAGUUGGAAAUAUGCACCACC----------------- .........((((.((((...(((((.((((.(((..(......)..))).))))...)))))))))....(....)..)))).....----------------- ( -24.00, z-score = 0.06, R) >droEre2.scaffold_4784 6089996 85 + 25762168 UUUUUUUUUUGCGAAAACCCGGAUGACCCCGACAG-CCCCGGUCGAUCCGGCGGGUUAUCACCACC--AGCCCCUCGCACACACCACA----------------- .........(((((....((((((((((.......-....)))).)))))).(((((.........--))))).))))).........----------------- ( -24.70, z-score = -2.27, R) >droYak2.chr3L 3947058 82 + 24197627 UUUUUUUUUUGCGAAAACCCGGAUGACCCCGACAG-ACCCGGUCGAUCCGGCGGGUUAUCACCACC--AGUUCCUCGCACACAUA-------------------- .........(((((.(((((((((((((.......-....)))).)))))).((.......))...--.)))..)))))......-------------------- ( -22.30, z-score = -1.44, R) >droSec1.super_2 3392586 93 + 7591821 ----CUUUUUGCGAAAACCCGGAUGACCCCGACAG-CCCCGGUCGAUCCGGCGGGUUAUCACCACC--AGCCCCUCGCACACACCACAUAC---ACAUGUACA-- ----.....(((((....((((((((((.......-....)))).)))))).(((((.........--))))).)))))......((((..---..))))...-- ( -25.30, z-score = -1.98, R) >droSim1.chr3L 2935468 93 + 22553184 ----CUUUUUGCGAAAACCCGGAUGACCCCGACAG-CCCCGGUCGAUCCGGCGGGUUAUCACCACC--AGCCCCUCGCACACACCACAUAC---AUAUGUACA-- ----.....(((((....((((((((((.......-....)))).)))))).(((((.........--))))).)))))......((((..---..))))...-- ( -25.50, z-score = -2.06, R) >consensus ____CUUUUUGCGAAAACCCGGAUGACCCCGACAG_CCCCGGUCGAUCCGGCGGGUUAUCACCACC__AGCCCCUCGCACACACCACA_________________ .........(((((...(((((((((((............)))).)))))).)(((....)))...........))))).......................... (-16.41 = -17.47 + 1.06)
| Location | 3,399,781 – 3,399,879 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 74.08 |
| Shannon entropy | 0.44661 |
| G+C content | 0.56896 |
| Mean single sequence MFE | -33.29 |
| Consensus MFE | -22.59 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.792927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3399781 98 - 24543557 CAUACACAUGUACAAUAUGUGGUGUGUGCGAGGGGCU--GGUGGUGAUAACCCGCCGGAUCGACCGGGG-CUGUCGGGGUCAUCCGGGUUUUCGCAAAAAG---- (((((((((((...)))))).)))))((((((((.((--((..(((((..((((((((.....))))..-....))))))))))))).)))))))).....---- ( -37.20, z-score = -1.41, R) >droAna3.scaffold_13337 13249409 88 + 23293914 -----------------GGUGGUGCAUAUUUCCAACUUGGCCAGUGAUAACCCAGCGGCUCGACCGGGGUCCGUUGGGGUCACUUUGGGCCUUGCAAAAAUCAAA -----------------(((..((((............((((((((((..(((((((((((.....))).))))))))))))))...)))).))))...)))... ( -35.52, z-score = -2.11, R) >droEre2.scaffold_4784 6089996 85 - 25762168 -----------------UGUGGUGUGUGCGAGGGGCU--GGUGGUGAUAACCCGCCGGAUCGACCGGGG-CUGUCGGGGUCAUCCGGGUUUUCGCAAAAAAAAAA -----------------.........((((((((.((--((..(((((..((((((((.....))))..-....))))))))))))).))))))))......... ( -30.00, z-score = -0.53, R) >droYak2.chr3L 3947058 82 - 24197627 --------------------UAUGUGUGCGAGGAACU--GGUGGUGAUAACCCGCCGGAUCGACCGGGU-CUGUCGGGGUCAUCCGGGUUUUCGCAAAAAAAAAA --------------------......((((((.((((--.(.((((((..((((.((((((.....)))-))).))))))))))).))))))))))......... ( -33.50, z-score = -2.86, R) >droSec1.super_2 3392586 93 - 7591821 --UGUACAUGU---GUAUGUGGUGUGUGCGAGGGGCU--GGUGGUGAUAACCCGCCGGAUCGACCGGGG-CUGUCGGGGUCAUCCGGGUUUUCGCAAAAAG---- --.((((....---))))........((((((((.((--((..(((((..((((((((.....))))..-....))))))))))))).)))))))).....---- ( -31.50, z-score = -0.07, R) >droSim1.chr3L 2935468 93 - 22553184 --UGUACAUAU---GUAUGUGGUGUGUGCGAGGGGCU--GGUGGUGAUAACCCGCCGGAUCGACCGGGG-CUGUCGGGGUCAUCCGGGUUUUCGCAAAAAG---- --...((((((---.......))))))(((((((.((--((..(((((..((((((((.....))))..-....))))))))))))).)))))))......---- ( -32.00, z-score = -0.35, R) >consensus _________________UGUGGUGUGUGCGAGGGGCU__GGUGGUGAUAACCCGCCGGAUCGACCGGGG_CUGUCGGGGUCAUCCGGGUUUUCGCAAAAAG____ ..........................((((((((.((.....((((((..((((.(((.((......)).))).)))))))))).)).))))))))......... (-22.59 = -22.85 + 0.26)
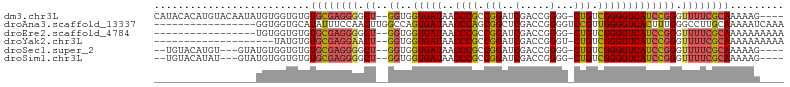

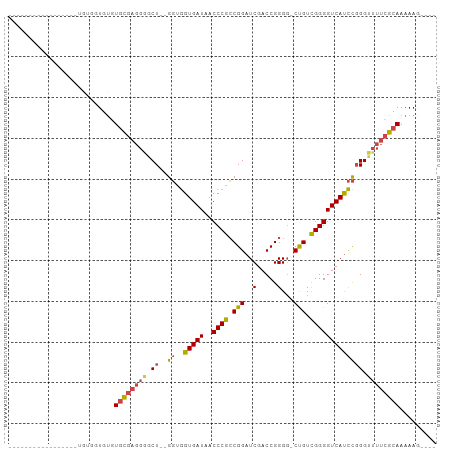
| Location | 3,399,837 – 3,399,928 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 73.36 |
| Shannon entropy | 0.37203 |
| G+C content | 0.49978 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -21.32 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.516980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3399837 91 - 24543557 ---GUAUGGAUGUGCCACUAGUGCUAUAUAGUGCUACAUACGUACAUACGCACAUACACAUGUACAAUAUGUGGUGUGUGCGAGGGGCUGGUGG ---...........((((((((.((.....((((.......))))...(((((((((((((((...)))))).))))))))).)).)))))))) ( -35.90, z-score = -1.48, R) >droEre2.scaffold_4784 6090056 75 - 25762168 GCAUCAUGGCUGUGCCACUAGUGCUAUGGAGCACUACAUACCUACGUG-------------------UGUGUGGUGUGUGCGAGGGGCUGGUGG ((......))....((((((((.((.((..(((((((((((......)-------------------))))))))))...)).)).)))))))) ( -31.20, z-score = -1.06, R) >droSim1.chr3L 2935524 86 - 22553184 ---GCAUGGAUGUGCCACUAGUGCUAUAUAGUACUACAUACAUACAUACAUCUGUACAUAUG-----UAUGUGGUGUGUGCGAGGGGCUGGUGG ---((((....))))(((((((.((.((((.(((((((((((((..(((....)))..))))-----)))))))))))))...)).))))))). ( -33.50, z-score = -1.64, R) >consensus ___GCAUGGAUGUGCCACUAGUGCUAUAUAGUACUACAUACAUACAUAC_____UACA_AUG_____UAUGUGGUGUGUGCGAGGGGCUGGUGG ..............((((((((.((.....((((((((((...........................))))))))))......)).)))))))) (-21.32 = -20.66 + -0.66)

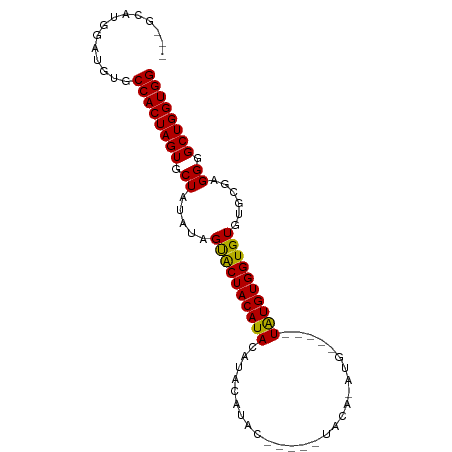
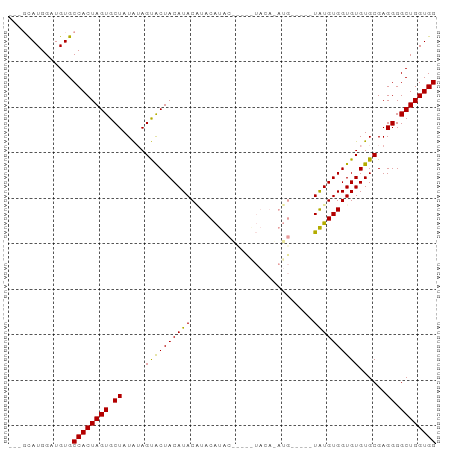
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:36 2011