| Sequence ID | dm3.chr3L |
|---|---|
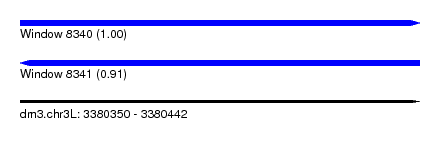
| Location | 3,380,350 – 3,380,442 |
| Length | 92 |
| Max. P | 0.998029 |

| Location | 3,380,350 – 3,380,442 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.85 |
| Shannon entropy | 0.51118 |
| G+C content | 0.53972 |
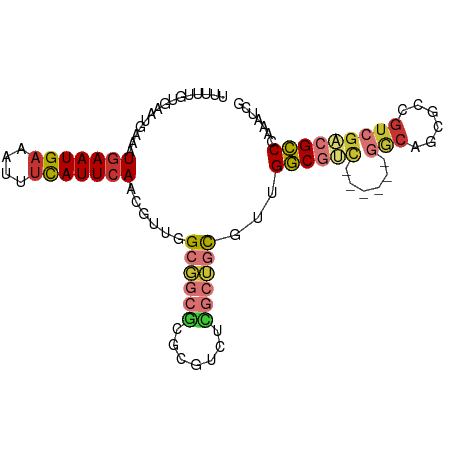
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -18.57 |
| Energy contribution | -18.76 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
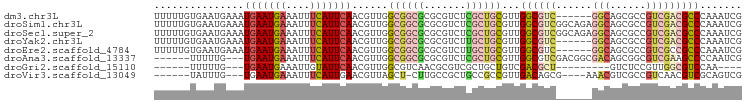
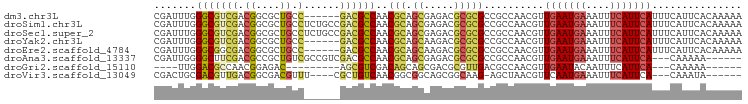
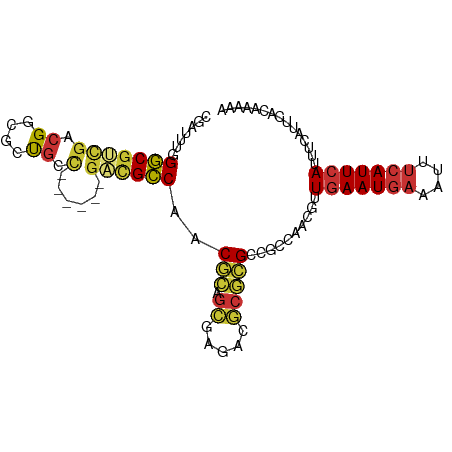
>dm3.chr3L 3380350 92 + 24543557 UUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUC------GGCAGCGCCGUCGACGCCCAAAUCG ..((((((((((((((.......)))))))))).((((((((((((..(((.((((.....))))..------))).))))))))))))..))))... ( -42.20, z-score = -3.55, R) >droSim1.chr3L 2915274 98 + 22553184 UUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGCAGAGGCAGCGCCGUCGACGCCCAAAUCG ..((((((((((((((.......)))))))))).((((((((((((..(((((((((((....)).)))).))))).))))))))))))..))))... ( -49.50, z-score = -4.75, R) >droSec1.super_2 3372861 98 + 7591821 UUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGCAGAGGCAGCGCCGUCGACGCCCAAAUCG ..((((((((((((((.......)))))))))).((((((((((((..(((((((((((....)).)))).))))).))))))))))))..))))... ( -49.50, z-score = -4.75, R) >droYak2.chr3L 3926962 92 + 24197627 UUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUUGCUGCGUUGGCGUC------GGCAGCGCCGUCGACGCCCAAAUCG ..((((((((((((((.......)))))))))).((((((((((((..(((.((((.....))))..------))).))))))))))))..))))... ( -40.40, z-score = -3.15, R) >droEre2.scaffold_4784 6070088 92 + 25762168 UUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUUGCUGCGUUGGCGUC------GGCAGCGCCGUCGCCGCCCAAAUCG ..((((((((((((((.......)))))))))).....(((((((((.((.((((((((....)).)------))))).)))).)))))))))))... ( -39.90, z-score = -2.83, R) >droAna3.scaffold_13337 13230375 89 - 23293914 ------UUUUUG---UGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGACGGCGACAGCGGCGUCGAAGCCCCAAUCG ------......---(((((((....)))))))..(((((.(((...((...((((((((((.((....)).)))).)))))).))..)))))))).. ( -38.00, z-score = -1.81, R) >droGri2.scaffold_15110 5784364 76 + 24565398 ------UUUUUG---UGAAUGAAAUUGUAUUCAACGUUGGCGUCAACGCGUCGCUGCUGUCGACGCU---------GUCUCCGUUGGCGUCCAA---- ------......---((((((......))))))...((((((((((((((((((....).)))))).---------(....)))))))).))))---- ( -24.90, z-score = -2.04, R) >droVir3.scaffold_13049 18170450 84 - 25233164 ------UAUUUG---UGAAUGAAAUUUCAUUGAACGUUAGCU-CUUGCCGCUGCCGCCGUUGACAGCG----AAACGUCGCCGUCAACGUCGCAGUCG ------.....(---(.(((((....)))))..))....((.-...)).(((((.(.(((((((.(((----(....)))).))))))).)))))).. ( -26.50, z-score = -1.74, R) >consensus UUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUC______GGCAGCGCCGUCGACGCCCAAAUCG ...............(((((((....))))))).((((((((.((((((...))).(((....)))...........))).))))))))......... (-18.57 = -18.76 + 0.19)
| Location | 3,380,350 – 3,380,442 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 73.85 |
| Shannon entropy | 0.51118 |
| G+C content | 0.53972 |
| Mean single sequence MFE | -32.31 |
| Consensus MFE | -13.85 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3380350 92 - 24543557 CGAUUUGGGCGUCGACGGCGCUGCC------GACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAA ...((((((((.((.((.((((((.------.........))))))...))))))))........((((((((((.......)))))))))))))).. ( -32.10, z-score = -1.47, R) >droSim1.chr3L 2915274 98 - 22553184 CGAUUUGGGCGUCGACGGCGCUGCCUCUGCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAA ...(((((((((.(.((((((.((.(((((.(.((....))).)).))).)))))))).).)))))(((((((((.......))))))))).)))).. ( -35.20, z-score = -1.88, R) >droSec1.super_2 3372861 98 - 7591821 CGAUUUGGGCGUCGACGGCGCUGCCUCUGCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAA ...(((((((((.(.((((((.((.(((((.(.((....))).)).))).)))))))).).)))))(((((((((.......))))))))).)))).. ( -35.20, z-score = -1.88, R) >droYak2.chr3L 3926962 92 - 24197627 CGAUUUGGGCGUCGACGGCGCUGCC------GACGCCAACGCAGCAAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAA ...(((((((((.(.((((((....------...((....)).((.....)))))))).).)))))(((((((((.......))))))))).)))).. ( -29.80, z-score = -1.11, R) >droEre2.scaffold_4784 6070088 92 - 25762168 CGAUUUGGGCGGCGACGGCGCUGCC------GACGCCAACGCAGCAAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAA ...(((((((((((..(((((....------).))))..(((........)))))))))).....((((((((((.......)))))))))))))).. ( -35.70, z-score = -2.71, R) >droAna3.scaffold_13337 13230375 89 + 23293914 CGAUUGGGGCUUCGACGCCGCUGUCGCCGUCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCA---CAAAAA------ ((.(((((((.((((((.((....)).)))))).)))..(((.(((...))))))...)))))).(((((((....)))))))---......------ ( -31.90, z-score = -1.01, R) >droGri2.scaffold_15110 5784364 76 - 24565398 ----UUGGACGCCAACGGAGAC---------AGCGUCGACAGCAGCGACGCGUUGACGCCAACGUUGAAUACAAUUUCAUUCA---CAAAAA------ ----((((.((.(((((....)---------.((((((.(....))))))))))).)))))).((.((((........)))))---).....------ ( -24.10, z-score = -2.85, R) >droVir3.scaffold_13049 18170450 84 + 25233164 CGACUGCGACGUUGACGGCGACGUUU----CGCUGUCAACGGCGGCAGCGGCAAG-AGCUAACGUUCAAUGAAAUUUCAUUCA---CAAAUA------ ((.((((..((((((((((((....)----)))))))))))...))))))....(-(((....))))(((((....)))))..---......------ ( -34.50, z-score = -4.08, R) >consensus CGAUUUGGGCGUCGACGGCGCUGCC______GACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAA .......((((((...(((...)))......))))))..(((.((.....)))))..........(((((((....)))))))............... (-13.85 = -14.27 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:33 2011