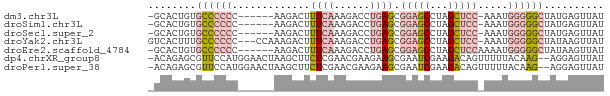
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,370,949 – 3,371,017 |
| Length | 68 |
| Max. P | 0.924049 |

| Location | 3,370,949 – 3,371,017 |
|---|---|
| Length | 68 |
| Sequences | 7 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 69.18 |
| Shannon entropy | 0.54402 |
| G+C content | 0.50247 |
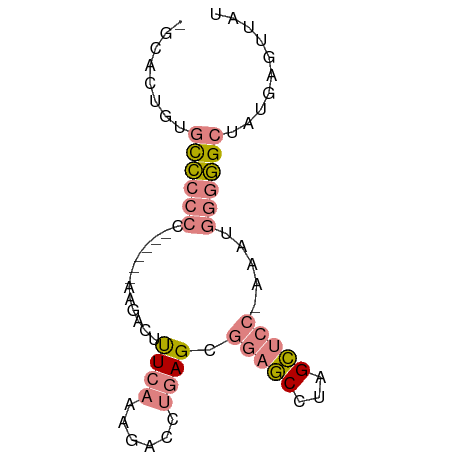
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -12.32 |
| Energy contribution | -13.19 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.924049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
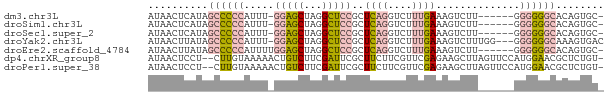
>dm3.chr3L 3370949 68 + 24543557 AUAACUCAUAGCCCCCAUUU-GGAGCUAGGCUCCGCUCAGGUCUUUGAAAGUCUU------GGGGGGCACAGUGC- ..........((((((....-(((((...)))))...((((.((.....)).)))------))))))).......- ( -26.50, z-score = -1.95, R) >droSim1.chr3L 2905864 68 + 22553184 AUAACUCAUAGCCCCCAUUU-GGAGCUAGGCUCCGCUCAGGUCUUUGAAAGUCUU------GGGGGGCACAGUGC- ..........((((((....-(((((...)))))...((((.((.....)).)))------))))))).......- ( -26.50, z-score = -1.95, R) >droSec1.super_2 3363459 68 + 7591821 AUAACUCAUAGCCCCCAUUU-GGAGCUAGGCUCCGCUCAGGUCUUUGAAAGUCUU------GGGGGGCACAGUGC- ..........((((((....-(((((...)))))...((((.((.....)).)))------))))))).......- ( -26.50, z-score = -1.95, R) >droYak2.chr3L 3917541 72 + 24197627 AUAACUUAUAGCCCCCAUUU-GGAGCUAGGCUCCGCUCAGGUCUUUGAAAGUCUUUGG---GGGGGGCAAAGUGAC ...((((...((((((....-(((((...))))).((((((.((.....)).).))))---))))))).))))... ( -26.80, z-score = -1.61, R) >droEre2.scaffold_4784 6060473 69 + 25762168 AUAACUUAUAGCCCCCAUUUUGGAGCUAGGCUCCGCUCAGGUCUUUGAAAGUCUU------GGGGGGCACAGUGC- ..........((((((.....(((((...)))))...((((.((.....)).)))------))))))).......- ( -26.50, z-score = -2.00, R) >dp4.chrXR_group8 5459705 73 + 9212921 AUAACUCCU--CUUGUAAAAACUGUCUUCGAUUCGCUUCUUCGUUCGAGAAGCUUAGUUCCAUGGAACGCUCUGU- .........--.......................(((((((.....)))))))...((((....)))).......- ( -12.90, z-score = -0.58, R) >droPer1.super_38 573873 73 - 801819 AUAACUCCU--CUUGUAAAAACUGUCUUCGAUUCGCUUCUUCGUUCGAGAAGCUUAGUUCCAUGGAACGCUCUGU- .........--.......................(((((((.....)))))))...((((....)))).......- ( -12.90, z-score = -0.58, R) >consensus AUAACUCAUAGCCCCCAUUU_GGAGCUAGGCUCCGCUCAGGUCUUUGAAAGUCUU______GGGGGGCACAGUGC_ ..........((((((.....(((((...)))))..((((....))))..............))))))........ (-12.32 = -13.19 + 0.86)
| Location | 3,370,949 – 3,371,017 |
|---|---|
| Length | 68 |
| Sequences | 7 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 69.18 |
| Shannon entropy | 0.54402 |
| G+C content | 0.50247 |
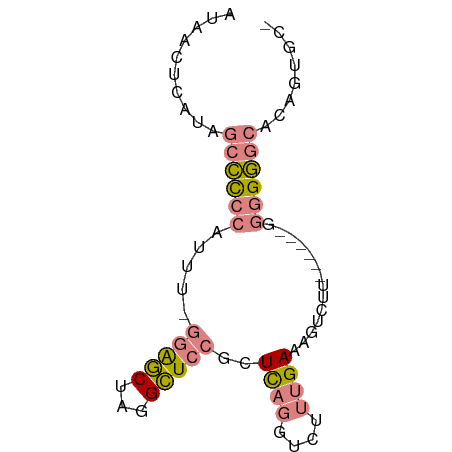
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -10.01 |
| Energy contribution | -11.36 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3370949 68 - 24543557 -GCACUGUGCCCCCC------AAGACUUUCAAAGACCUGAGCGGAGCCUAGCUCC-AAAUGGGGGCUAUGAGUUAU -.......((((((.------......((((......)))).(((((...)))))-....)))))).......... ( -21.90, z-score = -1.26, R) >droSim1.chr3L 2905864 68 - 22553184 -GCACUGUGCCCCCC------AAGACUUUCAAAGACCUGAGCGGAGCCUAGCUCC-AAAUGGGGGCUAUGAGUUAU -.......((((((.------......((((......)))).(((((...)))))-....)))))).......... ( -21.90, z-score = -1.26, R) >droSec1.super_2 3363459 68 - 7591821 -GCACUGUGCCCCCC------AAGACUUUCAAAGACCUGAGCGGAGCCUAGCUCC-AAAUGGGGGCUAUGAGUUAU -.......((((((.------......((((......)))).(((((...)))))-....)))))).......... ( -21.90, z-score = -1.26, R) >droYak2.chr3L 3917541 72 - 24197627 GUCACUUUGCCCCCC---CCAAAGACUUUCAAAGACCUGAGCGGAGCCUAGCUCC-AAAUGGGGGCUAUAAGUUAU ...((((.((((((.---.........((((......)))).(((((...)))))-....))))))...))))... ( -23.80, z-score = -2.35, R) >droEre2.scaffold_4784 6060473 69 - 25762168 -GCACUGUGCCCCCC------AAGACUUUCAAAGACCUGAGCGGAGCCUAGCUCCAAAAUGGGGGCUAUAAGUUAU -..(((..((((((.------......((((......)))).(((((...))))).....))))))....)))... ( -23.00, z-score = -1.90, R) >dp4.chrXR_group8 5459705 73 - 9212921 -ACAGAGCGUUCCAUGGAACUAAGCUUCUCGAACGAAGAAGCGAAUCGAAGACAGUUUUUACAAG--AGGAGUUAU -.......((((....))))..((((((((....(.((((((............)))))).)..)--))))))).. ( -15.70, z-score = -0.82, R) >droPer1.super_38 573873 73 + 801819 -ACAGAGCGUUCCAUGGAACUAAGCUUCUCGAACGAAGAAGCGAAUCGAAGACAGUUUUUACAAG--AGGAGUUAU -.......((((....))))..((((((((....(.((((((............)))))).)..)--))))))).. ( -15.70, z-score = -0.82, R) >consensus _GCACUGUGCCCCCC______AAGACUUUCAAAGACCUGAGCGGAGCCUAGCUCC_AAAUGGGGGCUAUGAGUUAU ........((((((.............((((......)))).(((((...))))).....)))))).......... (-10.01 = -11.36 + 1.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:30 2011