| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,326,156 – 3,326,246 |
| Length | 90 |
| Max. P | 0.544872 |

| Location | 3,326,156 – 3,326,246 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 65.64 |
| Shannon entropy | 0.64655 |
| G+C content | 0.39465 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
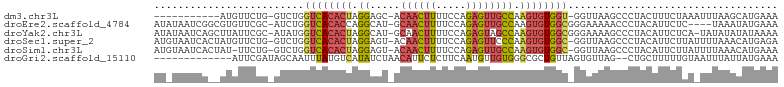
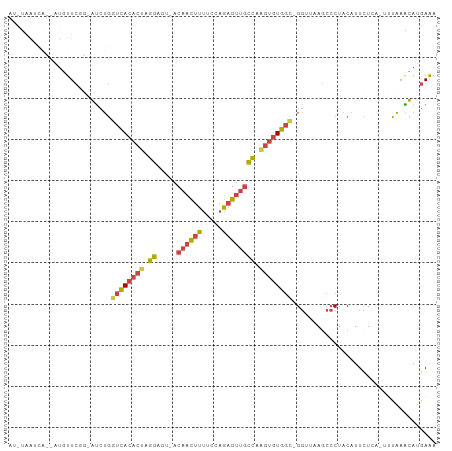
>dm3.chr3L 3326156 90 - 24543557 -----------AUGUUCUG-GUCUGGUCACACUAGGAGC-ACAACUUUUCCAGAGUUGCCAAGUGUGGU-GGUUAAGCCCUACUUUCUAAAUUUAAGCAUGAAA -----------(((((..(-((.(((.((((((.((...-.((((((.....)))))))).))))))((-((.......))))...))).)))..))))).... ( -20.30, z-score = 0.36, R) >droEre2.scaffold_4784 6015823 98 - 25762168 AUAUAAUCGGCGUGUUCGC-AUCUGGUCACACCAGGCAU-GCAACUUUUCCAGAGUUGCCAAGUGUGGCGGGAAAAACCCUACAUUCUC----UAAAUAUGAAA .........(((....)))-.(((((.....)))))(((-(((((((.....)))))))..(((((((.((......)))))))))...----.....)))... ( -26.20, z-score = -1.29, R) >droYak2.chr3L 3872841 101 - 24197627 AUAUAAUCAGCUUAUUCGC-AUAUGGUCACACUAGGCAU-GCAACUUUUCCAGAGUAGCCAAGUGUGGCGGGAAAAGCCCUACAUUCUCA-UAUAUAUAUAAAA (((((....((......))-(((((((((((((.(((.(-((............)))))).))))))))(((.....)))........))-))).))))).... ( -27.60, z-score = -2.89, R) >droSec1.super_2 3317824 101 - 7591821 AUGUAAUCACUAUGUUCUG-GUCUGGUCACACUAGGAGU-ACAACUUUUCCAGAGUUCCCAAGUGUGGC-GGUUAAGCCCUACAUUCUUAUUUUAAACAUGAGA ...........((((...(-((.(.((((((((.((.(.-..(((((.....)))))))).))))))))-.)....)))..))))((((((.......)))))) ( -21.00, z-score = 0.28, R) >droSim1.chr3L 2861363 100 - 22553184 AUGUAAUCACUAU-UUCUG-GUCUGGUCACACUAGGAGU-ACAACUUUUCCAGAGUUGCCAAGUGUGGC-GGUUAAGCCCUACAUUCUUAUUUUAAACAUGAAA ((((......(((-(((((-((........)))))))))-)((((((.....))))))...(((((((.-((.....)))))))))..........)))).... ( -22.40, z-score = -0.64, R) >droGri2.scaffold_15110 3279750 89 - 24565398 -------------AUUCGAUAGCAAUUUAUGUCAUAUCUAACAUUCUCUUCAAUGUUGUGGGCGCUGUUAGUGUUAG--CUGCUUUUUGUAAUUUAUUAUGAAA -------------...(((.((((......(((.....(((((((......)))))))..)))((((.......)))--)))))..)))............... ( -12.60, z-score = 0.74, R) >consensus AU_UAAUCA__AUGUUCGG_AUCUGGUCACACUAGGAGU_ACAACUUUUCCAGAGUUGCCAAGUGUGGC_GGUUAAGCCCUACAUUCUCA_UUUAAACAUGAAA ......................((.((((((((.((.....((((((.....)))))))).)))))))).))................................ (-11.00 = -11.87 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:24 2011