| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,287,551 – 3,287,707 |
| Length | 156 |
| Max. P | 0.731160 |

| Location | 3,287,551 – 3,287,707 |
|---|---|
| Length | 156 |
| Sequences | 6 |
| Columns | 158 |
| Reading direction | forward |
| Mean pairwise identity | 70.77 |
| Shannon entropy | 0.56244 |
| G+C content | 0.43565 |
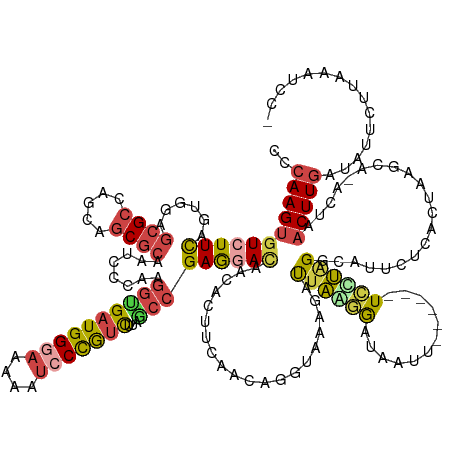
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -20.30 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
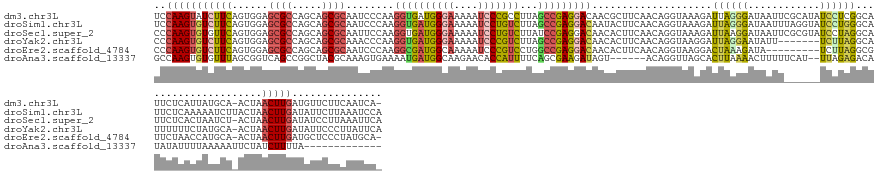
>dm3.chr3L 3287551 156 + 24543557 UCCAAGUAUCUUCAGUGGAGCGCCAGCAGCGCAAUCCCAAGGUGAUGGGAAAAAUCCCGCCUUAGCCGAGGACAACGCUUCAACAGGUAAAGAUUAGGGAUAAUUCGCAUAUCCUCGGCAUUCUCAUUAUGCA-ACUAACUUGAUGUUCUUCAAUCA- ..(((((.......((((.((((.....))))..(((((......)))))......))))....((((((((....(((......)))........(.((....)).)...))))))))..............-....)))))..............- ( -41.40, z-score = -0.91, R) >droSim1.chr3L 2822795 158 + 22553184 UCCAAGUGUCUUCAGUGGAGCGCCAGCAGCGCAAUCCCAAGGUGAUGGGAAAAAUCCUGUCUUAGCCGAGGACAAUACUUCAACAGGUAAAGAUUAGGGAUAAUUUAGGUAUCCUGGGCAUUCUCAAAAAUCUUACUAACUUGAUAUUCUUAAAUCCA ..(((((((((((.(.(((((((.....))))..))))..((((((((((....)))))))...)))))))))............(((((.((((.((((...((((((...))))))...))))...))))))))).)))))............... ( -43.10, z-score = -1.43, R) >droSec1.super_2 3280018 157 + 7591821 CCCAAGUGUGUUCAGUGGAGCGCCAGCAGCGCAAUUCCAAGGUGAUGGGAAAAAUCCUGUCUUAUCCGAGGACAACACUUCAACAGGUAAAGAUUAAGGAUAAUUCGCGUAUCCUAGGCAUUCUCACUAAUCU-ACUAACUUGAUAUCCUUAAAUUCA ...((((((((((..((((((((.....))))...)))).((.(((((((....)))))))....))..)))).))))))...(((((..((((((((((((.......)))))).((.....))..))))))-....)))))............... ( -39.40, z-score = -1.22, R) >droYak2.chr3L 3832868 150 + 24197627 CCCAAGUGUCUUCAGUGGAGCGCCAGCAGCGCAAACCCAAGGUGAUGGGAAAAAUCCCGUCUUAGCCGAGGACAACACUUCAACAGGUAAGGAUUAGGAAUAUU-------UCUUAGGCAUUUUUUCUAUGCA-ACUAACUUGAUAUUCCCUUAUUCA ......(((((((..(((.((((.....))))....))).((((((((((....)))))))...))))))))))................((((..((((((((-------..((((((((.......)))).-.))))...))))))))...)))). ( -44.70, z-score = -2.49, R) >droEre2.scaffold_4784 5970510 147 + 25762168 CCCAAGUGUCUUCAGUGGAGCGCCAGCAGCGCAAUCCCAAGGCGAUGGCAAAAAUCCCGUCCUGGCCGAGGACAACACUUCAACAGGUAAGGACUAAAGAUA---------UCUUAGGCGUUCUAACCAUGCA-ACUAACUUGAUGCUCCCUAUGCA- ..(((((.....((.(((((((((.((..(((.........)))...)).........(((((.(((((((......))))....))).)))))........---------.....)))))))))....))..-....))))).(((.......)))- ( -40.80, z-score = -0.46, R) >droAna3.scaffold_13337 7592936 137 + 23293914 GCCAAGUGUGUUUAGCGGUCAGCCGGCUACGCAAAGUGAAAAUGAUGGCAAGAACACCAUUUUCAGCGAAGAUAGU------ACAGGUUAGCACUUAAAACUUUUUCAU--UUAGAGACAUAUAUUUUAAAAAUUCUAUCUUUUA------------- ((....(((((..(((((....)).))))))))...((((((((.((.......)).))))))))))((((((((.------..((((....))))....((((.....--..))))..................))))))))..------------- ( -26.40, z-score = -0.19, R) >consensus CCCAAGUGUCUUCAGUGGAGCGCCAGCAGCGCAAUCCCAAGGUGAUGGGAAAAAUCCCGUCUUAGCCGAGGACAACACUUCAACAGGUAAAGAUUAAGGAUAAUU______UCCUAGGCAUUCUCACUAAGCA_ACUAACUUGAUAUUCUUAAAUCC_ ..(((((((((((......((((.....))))........((((((((((....)))))))...)))))))))....................((((((............)))))).....................)))))............... (-20.30 = -21.12 + 0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:18 2011