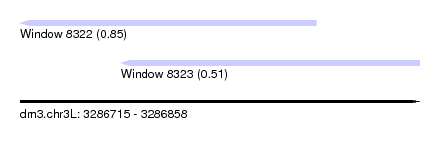
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,286,715 – 3,286,858 |
| Length | 143 |
| Max. P | 0.848315 |

| Location | 3,286,715 – 3,286,821 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.41 |
| Shannon entropy | 0.59030 |
| G+C content | 0.44721 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.49 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
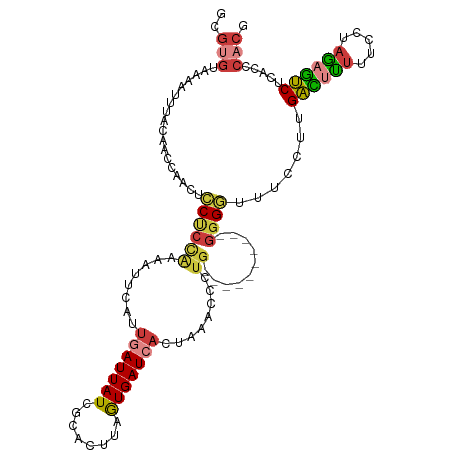
>dm3.chr3L 3286715 106 - 24543557 GCGUGUAAAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGCACCUAAUGAUCACUAAACCCUG----------GGGGUUUCCUUGACUUUUUCCUAGAGUCUCACCCACG .((((..................((((((........(((((((........)))))))........))----------)))).......((((((.....)))))).....)))) ( -20.39, z-score = -2.14, R) >droPer1.super_65 329838 108 + 397436 --------GCGUGGCGAAAAUUCCCCCUGAGGAUGUGGAAUUAUCGCCACCAAUGAUCGCCAAAUUCUGGCCAUGACUUGGGUCCUGGAGGGGCUUUUCUUUGCUCCUCACCCCCA --------(.(((((((.((((((((....))....)))))).)))))))).......((((.....)))).......((((.....(((((((........)))))))...)))) ( -41.20, z-score = -2.45, R) >droAna3.scaffold_13337 7592167 99 - 23293914 GCGUGUAAAAUUCUCCAACAUCCGCCCCGUAGGCGGUUUAUUAUCUCAUUAAAUGAUCACCCAA-----------------GUGUUCAAUGAAUUUUGGCCAAUUUCCCACCCUCA ..(((..(((((..((((...(((((.....))))).........((((....(((.(((....-----------------))).)))))))...))))..)))))..)))..... ( -21.90, z-score = -3.05, R) >droEre2.scaffold_4784 5969685 105 - 25762168 GCGUGUAAAAUUCCCAAGCAACUCCUCCAAAG-UCAUUGAUUAUCCCACUUAGUGAUCACAAAACCCUG----------GGGGUUUCCUUGACUCUUUCCUAGAGUCUCACCCACG .((((..........(((.....((((((..(-(...(((((((........)))))))....))..))----------))))....)))((((((.....)))))).....)))) ( -24.60, z-score = -2.60, R) >droYak2.chr3L 3832023 105 - 24197627 GCGUGUAAAAUUUGCAACCAACUCCUCCAAAC-UCAUUGAUUAUCACACUUAGUGAUCACAAAACCCUG----------GGGGUUUCCUUGACUUUUUCUUAGAGUCUCACCCACA ..(((..................((((((...-...(((...(((((.....)))))..))).....))----------)))).......((((((.....)))))).....))). ( -19.80, z-score = -0.94, R) >droSec1.super_2 3279197 106 - 7591821 GCGUGUAUAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGUGCUUAGUGAUCACUAAACCCUG----------GGGGUUUCCUUGACUUUUUCCUAGAGUCUCAGCCACG .((((..................((((((........(((((((........)))))))........))----------)))).......((((((.....)))))).....)))) ( -20.69, z-score = -1.07, R) >droSim1.chr3L 2821588 106 - 22553184 GCGUGUAUAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGUACUUAGUGAUCACUAAACCCUG----------GGGGUUUCCUUGACUUUUUCCUAGAGUCUCACCCACG .((((..................((((((........(((((((........)))))))........))----------)))).......((((((.....)))))).....)))) ( -20.69, z-score = -1.59, R) >consensus GCGUGUAAAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGCACUUAGUGAUCACUAAACCCUG__________GGGGUUUCCUUGACUUUUUCCUAGAGUCUCACCCACG .....................................(((((((........)))))))..(((((((...........)))))))....((((((.....))))))......... ( -8.82 = -9.49 + 0.67)

| Location | 3,286,751 – 3,286,858 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Shannon entropy | 0.42073 |
| G+C content | 0.40181 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -8.56 |
| Energy contribution | -8.51 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
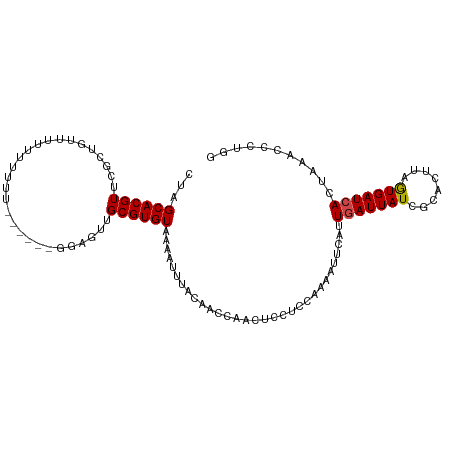
>dm3.chr3L 3286751 107 - 24543557 CUAGCACGUCCGCUGUUUUUUUUUUUUUUUUGGAGUUGCGUGUAAAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGCACCUAAUGAUCACUAAACCCUGG .((((......))))................(((((((..((((.....))))..))))))).(((........(((((((........)))))))........))) ( -20.39, z-score = -2.46, R) >droAna3.scaffold_13337 7592202 92 - 23293914 CAAGCACGUUUGGGGUUUUCGG---------GGAGUUGCGUGUAAAAUUCUCCAACAUCCGCCCCGUAGGCGGUUUAUUAUCUCAUUAAAUGAUCACCCAA------ ((((....))))((((..((((---------((((((........)))))))).....(((((.....)))))..................))..))))..------ ( -27.60, z-score = -2.01, R) >droEre2.scaffold_4784 5969721 97 - 25762168 CCAGCACGUUCGCUGUUUUUUG---------GGUGUUGCGUGUAAAAUUCCCAAGCAACUCCUCC-AAAGUCAUUGAUUAUCCCACUUAGUGAUCACAAAACCCUGG .((((......))))......(---------((((((((.((.........)).)))))......-........(((((((........)))))))....))))... ( -20.00, z-score = -0.99, R) >droYak2.chr3L 3832059 99 - 24197627 CUAGCACGUUUCCUGCUUUUUUUUU-------GUGUUGCGUGUAAAAUUUGCAACCAACUCCUCC-AAACUCAUUGAUUAUCACACUUAGUGAUCACAAAACCCUGG ..((((.......))))....((((-------((((((((.........))))))..........-.............(((((.....))))).))))))...... ( -13.80, z-score = -0.27, R) >droSec1.super_2 3279233 102 - 7591821 CUAGCACGUUCGGUGUUUUUUUUUUUU-----GAGUUGCGUGUAUAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGUGCUUAGUGAUCACUAAACCCUGG ...(((((.((((((..((((......-----((((((..((((.....))))..))))))....))))..))))))....)))))(((((....)))))....... ( -26.90, z-score = -4.14, R) >droSim1.chr3L 2821624 103 - 22553184 CUAGCACGUUUGCUAUUUUUUUUUUUU----GGAGUUGCGUGUAUAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGUACUUAGUGAUCACUAAACCCUGG .(((((....)))))............----(((((((..((((.....))))..))))))).(((........(((((((........)))))))........))) ( -22.39, z-score = -3.23, R) >consensus CUAGCACGUUCGCUGUUUUUUUUUU______GGAGUUGCGUGUAAAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGCACUUAGUGAUCACUAAACCCUGG ...((((((............................))))))...............................(((((((........)))))))........... ( -8.56 = -8.51 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:17 2011