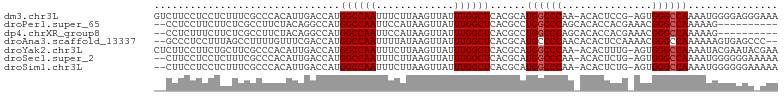
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,266,853 – 3,266,955 |
| Length | 102 |
| Max. P | 0.977265 |

| Location | 3,266,853 – 3,266,955 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.76 |
| Shannon entropy | 0.49610 |
| G+C content | 0.50095 |
| Mean single sequence MFE | -34.13 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
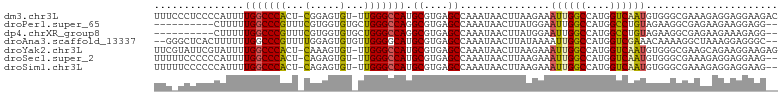
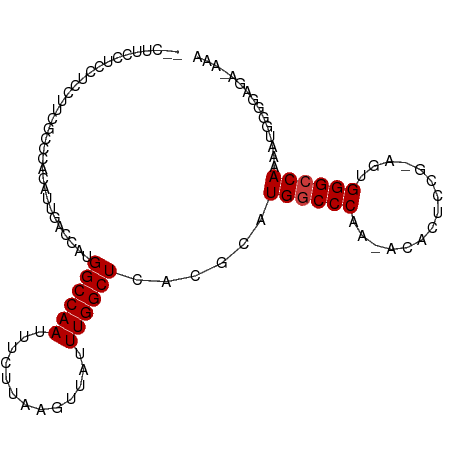
>dm3.chr3L 3266853 102 + 24543557 UUUCCCUCCCCAUUUUGGCCCACU-CGGAGUGU-UUGGGCCAUGCGUGAGCCAAAUAACUUAAGAAAUUGGCCAUGGUCAAUGUGGGCGAAAGAGGAGGAAGAC ....(((((...((((.((((((.-..((((..-...(((((....)).))).....)))).....((((((....)))))))))))).)))).)))))..... ( -37.80, z-score = -2.34, R) >droPer1.super_65 311077 92 - 397436 ----------CUUUUUGGCCCGUUUCGUGGUGUGCUGGGCCAGGCGUGAGCCAAAUAACUUAUGGAAUUGGCCAUGGCCUGUAGAAGGCGAGAAGAAGGAGG-- ----------((((((((((.((((((((((.......))).(((....))).........))))))).))))...((((.....)))))))))).......-- ( -33.90, z-score = -1.90, R) >dp4.chrXR_group8 5929663 92 - 9212921 ----------CUUUUUGGCCCGUUUCGUGGUGUGCUGGGCCAGGCGUGAGCCAAAUAACUUAUGGAAUUGGCCAUGGCCUGUAGAAGGCGAGAAGAAAGAGG-- ----------((((((((((.((((((((((.......))).(((....))).........))))))).))))...((((.....)))))))))).......-- ( -33.90, z-score = -2.16, R) >droAna3.scaffold_13337 7572510 100 + 23293914 --GGGCUCACUUUUUUGGCCCGUUUUGGAGUGUGUUGGGGCAUGCGUGAGCCAAAUAACUUAUAAAAUUGGCCAUGGUCGAAACAAAAGGCUAAAGGAGGGC-- --..((.(.(((((((((((.(((((((.((((((....))))))(((.(((((.............))))))))..)))))))....))))))))))))))-- ( -31.12, z-score = -1.28, R) >droYak2.chr3L 3812381 102 + 24197627 UUCGUAUUCGUAUUUUGGCCCACU-CAAAGUGU-UUGGGCCAUGCGUGAGCCAAAUAACUUAAGAAAUUGGCCAUGGUCAAUGUGGGCGAAGCAGAAGGAAGAG (((...(((((....(((((((..-(.....).-.))))))).((....))...............((((((....))))))....)))))...)))....... ( -29.80, z-score = -1.40, R) >droSec1.super_2 3260282 100 + 7591821 UUUUUCCCCCCAUUUUGGCCCACU-CAGAGUGU-UUGGGCCAUGCGUGAGCCAAAUAACUUAAGAAAUUGGCCAUGGUCAAUGUGGGCGAAAGAGGAGGAAG-- ...((((..((.((((.((((((.-..((((..-...(((((....)).))).....)))).....((((((....)))))))))))).)))).)).)))).-- ( -36.20, z-score = -2.49, R) >droSim1.chr3L 2802473 100 + 22553184 UUUUUCCCCCCAUUUUGGCCCACU-CAGAGUGU-UUGGGCCAUGCGUGAGCCAAAUAACUUAAGAAAUUGGCCAUGGUCAAUGUGGGCGAAAGAGGAGGAAG-- ...((((..((.((((.((((((.-..((((..-...(((((....)).))).....)))).....((((((....)))))))))))).)))).)).)))).-- ( -36.20, z-score = -2.49, R) >consensus UUU_UCUCCCCAUUUUGGCCCACU_CGGAGUGU_UUGGGCCAUGCGUGAGCCAAAUAACUUAAGAAAUUGGCCAUGGUCAAUGUGGGCGAAAAAGGAGGAAG__ ...............(((((((.............)))))))..((((.(((((.............)))))))))............................ (-19.40 = -19.30 + -0.10)
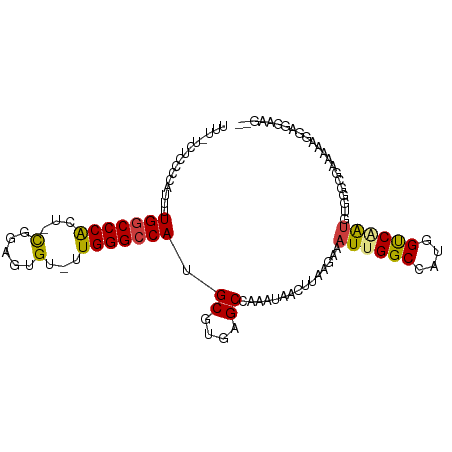
| Location | 3,266,853 – 3,266,955 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 73.76 |
| Shannon entropy | 0.49610 |
| G+C content | 0.50095 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3266853 102 - 24543557 GUCUUCCUCCUCUUUCGCCCACAUUGACCAUGGCCAAUUUCUUAAGUUAUUUGGCUCACGCAUGGCCCAA-ACACUCCG-AGUGGGCCAAAAUGGGGAGGGAAA ...((((((((((((.((((((.(((.((((((((((.............))))).....)))))..)))-.(.....)-.)))))).)))..))))))))).. ( -34.92, z-score = -2.06, R) >droPer1.super_65 311077 92 + 397436 --CCUCCUUCUUCUCGCCUUCUACAGGCCAUGGCCAAUUCCAUAAGUUAUUUGGCUCACGCCUGGCCCAGCACACCACGAAACGGGCCAAAAAG---------- --......................((((...((((((.............))))))...))))(((((...............)))))......---------- ( -22.28, z-score = -1.17, R) >dp4.chrXR_group8 5929663 92 + 9212921 --CCUCUUUCUUCUCGCCUUCUACAGGCCAUGGCCAAUUCCAUAAGUUAUUUGGCUCACGCCUGGCCCAGCACACCACGAAACGGGCCAAAAAG---------- --......................((((...((((((.............))))))...))))(((((...............)))))......---------- ( -22.28, z-score = -1.25, R) >droAna3.scaffold_13337 7572510 100 - 23293914 --GCCCUCCUUUAGCCUUUUGUUUCGACCAUGGCCAAUUUUAUAAGUUAUUUGGCUCACGCAUGCCCCAACACACUCCAAAACGGGCCAAAAAAGUGAGCCC-- --...........(((...((.......)).)))..................(((((((....((((................)))).......))))))).-- ( -20.09, z-score = -0.49, R) >droYak2.chr3L 3812381 102 - 24197627 CUCUUCCUUCUGCUUCGCCCACAUUGACCAUGGCCAAUUUCUUAAGUUAUUUGGCUCACGCAUGGCCCAA-ACACUUUG-AGUGGGCCAAAAUACGAAUACGAA .............((((.......((.....((((((.............))))))....))(((((((.-.(.....)-..))))))).....))))...... ( -21.62, z-score = -0.58, R) >droSec1.super_2 3260282 100 - 7591821 --CUUCCUCCUCUUUCGCCCACAUUGACCAUGGCCAAUUUCUUAAGUUAUUUGGCUCACGCAUGGCCCAA-ACACUCUG-AGUGGGCCAAAAUGGGGGGAAAAA --.(((((((..(((.((((((.(((.((((((((((.............))))).....)))))..)))-.(.....)-.)))))).)))..))))))).... ( -33.02, z-score = -2.09, R) >droSim1.chr3L 2802473 100 - 22553184 --CUUCCUCCUCUUUCGCCCACAUUGACCAUGGCCAAUUUCUUAAGUUAUUUGGCUCACGCAUGGCCCAA-ACACUCUG-AGUGGGCCAAAAUGGGGGGAAAAA --.(((((((..(((.((((((.(((.((((((((((.............))))).....)))))..)))-.(.....)-.)))))).)))..))))))).... ( -33.02, z-score = -2.09, R) >consensus __CUUCCUCCUCCUUCGCCCACAUUGACCAUGGCCAAUUUCUUAAGUUAUUUGGCUCACGCAUGGCCCAA_ACACUCCG_AGUGGGCCAAAAUGGGGAGA_AAA ...............................((((((.............))))))......((((((...............))))))............... (-15.49 = -15.64 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:16 2011