
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,252,509 – 3,252,613 |
| Length | 104 |
| Max. P | 0.500000 |

| Location | 3,252,509 – 3,252,613 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 63.47 |
| Shannon entropy | 0.70831 |
| G+C content | 0.39103 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -9.75 |
| Energy contribution | -10.31 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.76 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
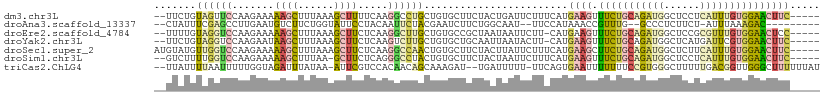
Download alignment: ClustalW | MAF
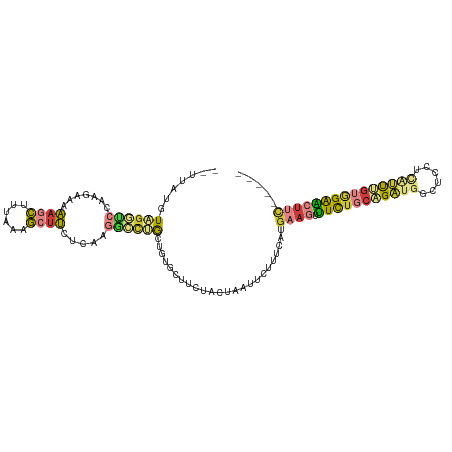
>dm3.chr3L 3252509 104 + 24543557 --UUCUGUAGUUCCAAGAAAAAGCUUUAAAGCUUUUCAAGGCCUGCUGUGCUUCUACUGAUUCUUUCAUGAAGUUUCUGCAGAUGGCUCCUCAUUUGUGGAACUUC----- --......(((((((.(((((.(((....))))))))..((((..(((((((((...(((.....))).)))))....))))..)))).........)))))))..----- ( -28.80, z-score = -1.85, R) >droAna3.scaffold_13337 7557772 95 + 23293914 --CUAUUUCGAGCCUUGAAUGUCUUCUGGUAUUCCUACAAUUCUACGAAUCUUCUGGCAAU--UUCCAUAAACCGUUUG--GCCCUCUUCU-AUUUAAAGAC--------- --.......(((..(..((((..((.(((.(((((....((((...)))).....)).)))--..))).))..))))..--)..)))....-..........--------- ( -12.60, z-score = -0.00, R) >droEre2.scaffold_4784 5935003 103 + 25762168 --UUUUGUAGGUCCAAGAAAAAGCUUUAAAGCUUCUCAAGGCUUGCUGUGCCGCUAAUAAUUCUU-CAUGAAGUUUCUGCAGAUGGCUCCGCGUUUGUGGAACUCC----- --.((((((((..(..((..(((((....))))).))..(((.......))).............-......)..)))))))).((.(((((....))))).))..----- ( -23.20, z-score = 0.38, R) >droYak2.chr3L 3797175 103 + 24197627 --UUCUGUAGGUCCAAGAAUAAGCUUUAAAGCUUCUCAAGUCUUGCUGUGCUGCAAUUAAUACUU-CAUGAAGUUUCUGCAGAUGGCUCAUGAUUCGUGGAACUUC----- --......(((((((.(((((((((....)))))..........(((((.(((((......((((-....))))...)))))))))).....)))).))).)))).----- ( -23.30, z-score = -0.23, R) >droSec1.super_2 3246039 106 + 7591821 AUGUAUGUUGGUCCAAGAAAAAGCUUUAAAGCUUCUCAAGGCCAACUGUGCUUCUACUUAUUCUUUCAUGAAGCUUCUGCAGAUGGCUCUUCAUUUGUGGAACUUC----- ..(((((((((((..(((...((((....)))))))...))))))).))))..................((((.(((..(((((((....)))))))..)))))))----- ( -30.60, z-score = -2.72, R) >droSim1.chr3L 2785916 103 + 22553184 --GUCUUUUGGUCCAAGAAAAAGCUUUAA-GCUUCUCAGGGCCUACUGUGCUUCUACUAAUUCUUUCAUGAAGUUUCUGCAGAUGGCUCCUCAUUUGUGGAACUUC----- --.......(((((.(((...(((.....-))))))..)))))..........................((((((.(..(((((((....)))))))..)))))))----- ( -25.60, z-score = -1.12, R) >triCas2.ChLG4 12562593 105 + 13894384 --UUAUUUUAAUUUUUGGUAGAUUUAUAA-AUUCGUCCACAACAGCAAAGAU--UGAUUUUU-UUCAGUGAAUUUUUUUCCGUGGGCUUUUUGACGGUUGGGCUUUUUUAU --....((((((..(..(..((.......-..))((((((..........((--(((.....-.)))))(((.....))).))))))...)..)..))))))......... ( -14.30, z-score = 0.80, R) >consensus __UUAUGUAGGUCCAAGAAAAAGCUUUAAAGCUUCUCAAGGCCUGCUGUGCUUCUACUAAUUCUUUCAUGAAGUUUCUGCAGAUGGCUCCUCAUUUGUGGAACUUC_____ .......((((((.......((((......)))).....))))))........................((((.(((((((((((......)))))))))))))))..... ( -9.75 = -10.31 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:12 2011