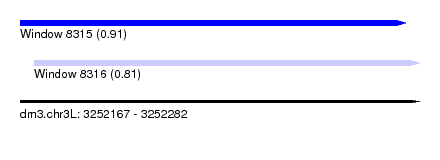
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,252,167 – 3,252,282 |
| Length | 115 |
| Max. P | 0.909141 |

| Location | 3,252,167 – 3,252,278 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.13 |
| Shannon entropy | 0.38415 |
| G+C content | 0.29566 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -14.75 |
| Energy contribution | -16.03 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.909141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
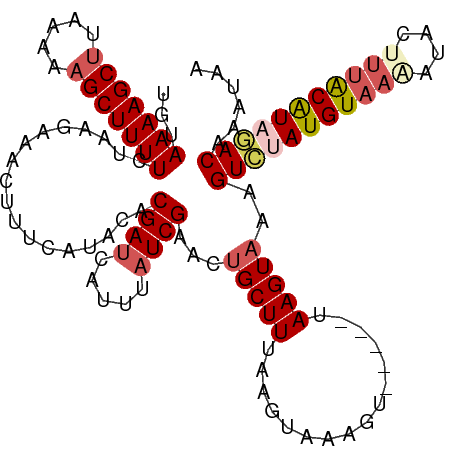
>dm3.chr3L 3252167 111 + 24543557 UGUAAAAGCUUGAACAGCUUUUCUAAGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGU-----UAAGUAAAGUAAAUGUAAGAUACUUUACAUAGACAAAAAA ....((((((.....))))))((((...((((((......((((.....)))).(((......))))))))-----)..((((((((.........)))))))).))))....... ( -17.90, z-score = -0.75, R) >droSim1.chr3L 2785570 116 + 22553184 UGUAAAAGCUUAAAAAGCUUUUCUAGGGAACUUUCAUACACGAUCAUUUUUCGAACUGCUUUAAGUAAGGUAAGGUUAAGUAAAGUCGAUGUAAAAUACUUUACAUAGACAAAUAA ...(((((((.....)))))))..((....))....(((.(((.......)))(((((((((....)))))..))))..)))..(((.(((((((....))))))).)))...... ( -19.80, z-score = -0.66, R) >droSec1.super_2 3245713 116 + 7591821 UGUAAAAGCUUAAAAAGCUUUUCUAGGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGUAAGGUUAAGUAAGGUAUAUGUAAAAUACUUUACAUAUACAAAUAA ((((((((((.....))))))...((....))....))))......(((((..(((((((((....)))))..))))..)))))(((((((((((....)))))))))))...... ( -24.50, z-score = -3.00, R) >droYak2.chr3L 3796828 109 + 24197627 UGAAAAAGCUUAAAGAGCUUUUCUAACCUACAUCAGCACACGAUCAUUUAUCGAACUGCUUUACG--AAUU-----UAAGUUAAGUCUAUGUAAAAUACCUUGCGUAGACAGCUUA ...((((((((...))))))))............((((..((((.....))))...)))).....--....-----((((((..((((((((((......)))))))))))))))) ( -24.20, z-score = -2.57, R) >droEre2.scaffold_4784 5934682 101 + 25762168 UUAAAAAGCAAGGAGAGCUUUUUUAAACUACAU---CACACGAUCAUUUAUCGAACUGCUUUACG--AACU-----UAAGUAAAGUCUAUGUAAAAUACAG-ACGUAGACUA---- ..(((((((.......)))))))....((((.(---(...((((.....))))....(((((((.--....-----...)))))))..............)-).))))....---- ( -18.50, z-score = -1.65, R) >consensus UGUAAAAGCUUAAAAAGCUUUUCUAAGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGU_____UAAGUAAAGUCUAUGUAAAAUACUUUACAUAGACAAAUAA ...(((((((.....)))))))..................((((.....))))...(((((................)))))..(((((((((((....)))))))))))...... (-14.75 = -16.03 + 1.28)

| Location | 3,252,171 – 3,252,282 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Shannon entropy | 0.38205 |
| G+C content | 0.30007 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -13.37 |
| Energy contribution | -14.65 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
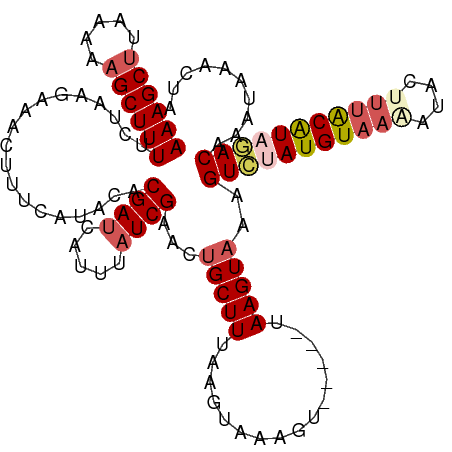
>dm3.chr3L 3252171 111 + 24543557 AAAGCUUGAACAGCUUUUCUAAGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGU-----UAAGUAAAGUAAAUGUAAGAUACUUUACAUAGACAAAAAAACUA ((((((.....))))))((((...((((((......((((.....)))).(((......))))))))-----)..((((((((.........)))))))).))))........... ( -17.60, z-score = -0.95, R) >droSim1.chr3L 2785574 116 + 22553184 AAAGCUUAAAAAGCUUUUCUAGGGAACUUUCAUACACGAUCAUUUUUCGAACUGCUUUAAGUAAGGUAAGGUUAAGUAAAGUCGAUGUAAAAUACUUUACAUAGACAAAUAAACUA ...((((((...(((((.((((....))........(((.......)))..........)).)))))....))))))...(((.(((((((....))))))).))).......... ( -18.80, z-score = -0.49, R) >droSec1.super_2 3245717 115 + 7591821 AAAGCUUAAAAAGCUUUUCUAGGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGUAAGGUUAAGUAAGGUAUAUGUAAAAUACUUUACAUAUACAAAUAAACU- ...((((((...(((((.((((....))........((((.....))))..........)).)))))....))))))...(((((((((((....))))))))))).........- ( -23.10, z-score = -2.90, R) >droYak2.chr3L 3796832 109 + 24197627 AAAGCUUAAAGAGCUUUUCUAACCUACAUCAGCACACGAUCAUUUAUCGAACUGCUUUACG--AAUU-----UAAGUUAAGUCUAUGUAAAAUACCUUGCGUAGACAGCUUAACUU (((((((...))))))).............((((..((((.....))))...)))).....--...(-----((((((..((((((((((......)))))))))))))))))... ( -24.40, z-score = -2.93, R) >droEre2.scaffold_4784 5934686 101 + 25762168 AAAGCAAGGAGAGCUUUUUUAAACUACAUC---ACACGAUCAUUUAUCGAACUGCUUUACG--AACU-----UAAGUAAAGUCUAUGUAAAAUACAG-ACGUAGAC----UAACUC (((((.......)))))......((((.((---...((((.....))))....(((((((.--....-----...)))))))..............)-).))))..----...... ( -16.50, z-score = -0.74, R) >consensus AAAGCUUAAAAAGCUUUUCUAAGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGU_____UAAGUAAAGUCUAUGUAAAAUACUUUACAUAGACAAAUAAACUA ((((((.....))))))...................((((.....))))...(((((................)))))..(((((((((((....))))))))))).......... (-13.37 = -14.65 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:11 2011