| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,251,024 – 3,251,104 |
| Length | 80 |
| Max. P | 0.999242 |

| Location | 3,251,024 – 3,251,104 |
|---|---|
| Length | 80 |
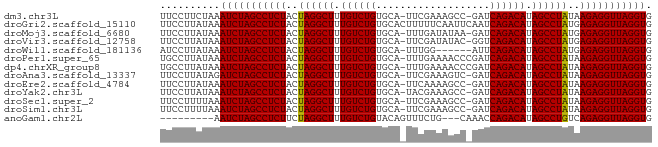
| Sequences | 13 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 88.54 |
| Shannon entropy | 0.27652 |
| G+C content | 0.42021 |
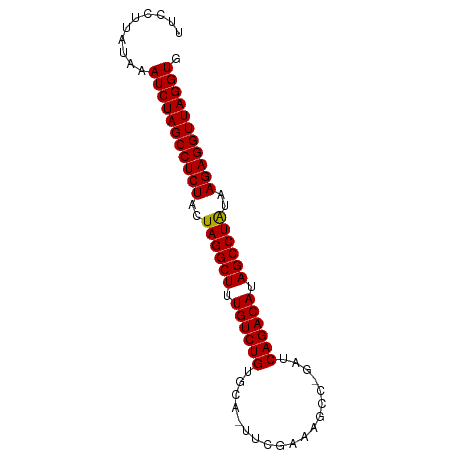
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -32.89 |
| Energy contribution | -32.82 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.04 |
| Mean z-score | -6.36 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3251024 80 + 24543557 UUCCUUCUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUCGAAAGCC-GAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-.(((.....)-)).)))))).))))))..))))))))))). ( -35.70, z-score = -6.46, R) >droGri2.scaffold_15110 607073 82 + 24565398 UUCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCACUUUUUCAAUUCAAUCAGACAUAGCCUAUGAGAGGUUAGGUG ..........(((((((((((..((((((.((((((...................)))))).))))))..))))))))))). ( -33.31, z-score = -6.50, R) >droMoj3.scaffold_6680 22800806 80 - 24764193 UUCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUUGAUAUAA-GAUCAGACAUAGCCUAUGAGAGGUUAGGUG ..........(((((((((((..((((((.((((((...(-(((......)-))))))))).))))))..))))))))))). ( -34.30, z-score = -5.92, R) >droVir3.scaffold_12758 61327 80 - 972128 UUCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUCGAUAUAC-GGUCAGACAUAGCCUAUGAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-.(((.....)-)).)))))).))))))..))))))))))). ( -35.10, z-score = -5.90, R) >droWil1.scaffold_181136 1999691 75 + 2313701 AUCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUUGG------AUUCAGACAUAGCCUAUGAGAGGUUAGGUG ..........(((((((((((..((((((.((((((..(.-....)------...)))))).))))))..))))))))))). ( -34.30, z-score = -6.04, R) >droPer1.super_65 293396 81 - 397436 UGCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUUGAAAACCCGAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-.(((......))).)))))).))))))..))))))))))). ( -33.80, z-score = -6.19, R) >dp4.chrXR_group8 5912838 81 - 9212921 UGCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUUGAAAACCCGAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-.(((......))).)))))).))))))..))))))))))). ( -33.80, z-score = -6.19, R) >droAna3.scaffold_13337 7556609 80 + 23293914 UUCCUUAUAGAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUCGAAAGUC-GAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-.(((.....)-)).)))))).))))))..))))))))))). ( -36.40, z-score = -6.38, R) >droEre2.scaffold_4784 5933725 80 + 25762168 UUCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUCAAAAGCC-GAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((.((.-.......)).-...)))))).))))))..))))))))))). ( -35.00, z-score = -6.79, R) >droYak2.chr3L 3795941 80 + 24197627 UUCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UACGAAAGCC-GAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-..(....)..-...)))))).))))))..))))))))))). ( -35.50, z-score = -6.50, R) >droSec1.super_2 3244762 80 + 7591821 UUCCUUUUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUCGAAAGCC-GAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-.(((.....)-)).)))))).))))))..))))))))))). ( -35.70, z-score = -6.66, R) >droSim1.chr3L 2784398 80 + 22553184 UUCCUUUUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA-UUCGAAAGCC-GAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((....-.(((.....)-)).)))))).))))))..))))))))))). ( -35.70, z-score = -6.66, R) >anoGam1.chr2L 31868088 70 - 48795086 ---------AAUCUAGCCUCUUCUAGGCUUUGUCUGUACAGUUUCUG---CAAACCAGACAUAGCCUGUCAGAGGUUAGGUG ---------.(((((((((((..((((((.((((((....((....)---)....)))))).))))))..))))))))))). ( -34.00, z-score = -6.45, R) >consensus UUCCUUAUAAAUCUAGCCUCUACUAGGCUUUGUCUGUGCA_UUCGAAAGCC_GAUCAGACAUAGCCUAUAAGAGGUUAGGUG ..........(((((((((((..((((((.((((((...................)))))).))))))..))))))))))). (-32.89 = -32.82 + -0.07)
| Location | 3,251,024 – 3,251,104 |
|---|---|
| Length | 80 |
| Sequences | 13 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 88.54 |
| Shannon entropy | 0.27652 |
| G+C content | 0.42021 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -26.36 |
| Energy contribution | -26.43 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.58 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3251024 80 - 24543557 CACCUAACCUCUUAUAGGCUAUGUCUGAUC-GGCUUUCGAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAGAAGGAA ...(((.(((((..((((((.((((((...-.((.......-.)).)))))).))))))..))))).)))............ ( -30.10, z-score = -4.31, R) >droGri2.scaffold_15110 607073 82 - 24565398 CACCUAACCUCUCAUAGGCUAUGUCUGAUUGAAUUGAAAAAGUGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGAA ...(((.(((((..((((((.((((((..((..((....))...)))))))).))))))..))))).)))............ ( -29.00, z-score = -4.87, R) >droMoj3.scaffold_6680 22800806 80 + 24764193 CACCUAACCUCUCAUAGGCUAUGUCUGAUC-UUAUAUCAAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGAA ...(((.(((((..((((((.((((((...-..........-....)))))).))))))..))))).)))............ ( -26.83, z-score = -4.32, R) >droVir3.scaffold_12758 61327 80 + 972128 CACCUAACCUCUCAUAGGCUAUGUCUGACC-GUAUAUCGAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGAA ...(((.(((((..((((((.((((((...-((((.....)-))).)))))).))))))..))))).)))............ ( -29.60, z-score = -5.08, R) >droWil1.scaffold_181136 1999691 75 - 2313701 CACCUAACCUCUCAUAGGCUAUGUCUGAAU------CCAAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGAU ...(((.(((((..((((((.((((((...------.....-....)))))).))))))..))))).)))............ ( -27.20, z-score = -4.87, R) >droPer1.super_65 293396 81 + 397436 CACCUAACCUCUUAUAGGCUAUGUCUGAUCGGGUUUUCAAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGCA ...(((.(((((..((((((.((((((...(.(((....))-).).)))))).))))))..))))).)))............ ( -27.80, z-score = -3.78, R) >dp4.chrXR_group8 5912838 81 + 9212921 CACCUAACCUCUUAUAGGCUAUGUCUGAUCGGGUUUUCAAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGCA ...(((.(((((..((((((.((((((...(.(((....))-).).)))))).))))))..))))).)))............ ( -27.80, z-score = -3.78, R) >droAna3.scaffold_13337 7556609 80 - 23293914 CACCUAACCUCUUAUAGGCUAUGUCUGAUC-GACUUUCGAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUCUAUAAGGAA ...(((.(((((..((((((.((((((.((-(.....))).-....)))))).))))))..))))).)))............ ( -29.10, z-score = -4.62, R) >droEre2.scaffold_4784 5933725 80 - 25762168 CACCUAACCUCUUAUAGGCUAUGUCUGAUC-GGCUUUUGAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGAA ...(((.(((((..((((((.((((((...-.((.......-.)).)))))).))))))..))))).)))............ ( -30.10, z-score = -4.69, R) >droYak2.chr3L 3795941 80 - 24197627 CACCUAACCUCUUAUAGGCUAUGUCUGAUC-GGCUUUCGUA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGAA ...(((.(((((..((((((.((((((...-.((.......-.)).)))))).))))))..))))).)))............ ( -30.10, z-score = -4.41, R) >droSec1.super_2 3244762 80 - 7591821 CACCUAACCUCUUAUAGGCUAUGUCUGAUC-GGCUUUCGAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAAAAGGAA ...(((.(((((..((((((.((((((...-.((.......-.)).)))))).))))))..))))).)))............ ( -30.10, z-score = -4.74, R) >droSim1.chr3L 2784398 80 - 22553184 CACCUAACCUCUUAUAGGCUAUGUCUGAUC-GGCUUUCGAA-UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAAAAGGAA ...(((.(((((..((((((.((((((...-.((.......-.)).)))))).))))))..))))).)))............ ( -30.10, z-score = -4.74, R) >anoGam1.chr2L 31868088 70 + 48795086 CACCUAACCUCUGACAGGCUAUGUCUGGUUUG---CAGAAACUGUACAGACAAAGCCUAGAAGAGGCUAGAUU--------- ...(((.(((((...(((((.((((((...((---(((...))))))))))).)))))...))))).)))...--------- ( -29.70, z-score = -5.26, R) >consensus CACCUAACCUCUUAUAGGCUAUGUCUGAUC_GGCUUUCGAA_UGCACAGACAAAGCCUAGUAGAGGCUAGAUUUAUAAGGAA ...(((.(((((..((((((.((((((...................)))))).))))))..))))).)))............ (-26.36 = -26.43 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:10 2011