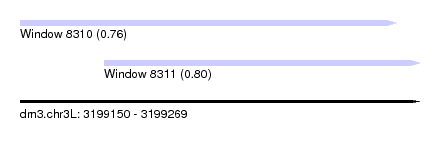
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,199,150 – 3,199,269 |
| Length | 119 |
| Max. P | 0.797525 |

| Location | 3,199,150 – 3,199,262 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.73 |
| Shannon entropy | 0.71917 |
| G+C content | 0.28247 |
| Mean single sequence MFE | -18.21 |
| Consensus MFE | -7.29 |
| Energy contribution | -6.77 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
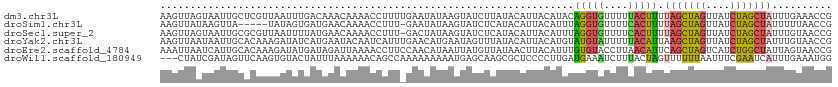
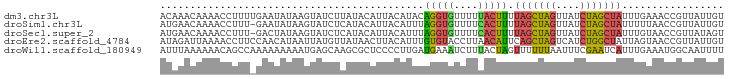
>dm3.chr3L 3199150 112 + 24543557 AAGUUAGUAAUUGCUCGUUAAUUUGACAAACAAAACCUUUUGAAUAUAAGUAUCUUAUACAUUACAUACAGGUGUUUUUACUUUUAGCUAGUUAUCUAGCUAUUUGAAACCG .....(((((..((..(((.....))).......((((..((((((((((...))))))..)).))...))))))..)))))..(((((((....))))))).......... ( -17.40, z-score = -0.55, R) >droSim1.chr3L 2729263 106 + 22553184 AAGUUAUAAGUUA-----UAUAGUGAUGAACAAAACCUUU-GAAUAUAAGUAUCUCAUACAUUACAUUUAGGUGUUUUCACUUUUAGCUAGUUAUCUAGCUAUUUUUAACCG ..((((.(((...-----...(((((.((((...((((..-((((.(((((((...)))).))).)))))))))))))))))..(((((((....)))))))))).)))).. ( -20.10, z-score = -1.99, R) >droSec1.super_2 3192291 111 + 7591821 AAGUUAGUAAUUGCGCGUUAAUUUUAUGAACAAAACCUUU-GACUAUAAGUAUCUCAUACAUUACAUUUAGGUGUUUUCACUUUUAGCUAGUUAUCUAGCUAUUUGUAACCG .((((((...(((..(((.......)))..))).....))-))))....((((...)))).(((((...(((((....))))).(((((((....)))))))..)))))... ( -18.50, z-score = -0.74, R) >droYak2.chr3L 3744487 112 + 24197627 AAGUUAAUAAUUGCACAAAGAUAUCAUGAAUACAAUCAUUUGAACAUGAAUAGUUUAUACAUUACAUGUAUGUAUUUUUACAUUAAGCUAGUUAUCUAGCUAUUUGUAACCG ...........................(((((((.((....))((((((((.((....))))).))))).)))))))(((((...((((((....))))))...)))))... ( -19.10, z-score = -1.05, R) >droEre2.scaffold_4784 5881811 112 + 25762168 AAAUUAAUCAUUGCACAAAGAUAUGAUAGAUUAAAACCUUCCAACAUAAUUAUGUUAUAACUUACAUUUGUGUACCUUAACAUUCAGCUAGUCAUCUGGCUAUUAGUAACCG ..((((((...(((((((((.((((((((((((.............))))).))))))).)).....)))))))...........((((((....))))))))))))..... ( -17.12, z-score = -1.33, R) >droWil1.scaffold_180949 5025722 109 + 6375548 ---CUAUCGAUAGUUCAAGUGUACUAUUUAAAAAACAGCCAAAAAAAAAUGAGCAAGCGCUCCCCUUGAUGAAAUCUUUACUAGUUUUUUAAUUUCGAAUCAUUUGAAAUGG ---..........((((((((......(((((((((..............((((....)))).....(((...))).......)))))))))........)))))))).... ( -17.04, z-score = -0.90, R) >consensus AAGUUAAUAAUUGCACAAUAAUAUGAUGAACAAAACCUUU_GAAUAUAAGUAUCUUAUACAUUACAUUUAGGUAUUUUUACUUUUAGCUAGUUAUCUAGCUAUUUGUAACCG .....................................................................(((((....))))).(((((((....))))))).......... ( -7.29 = -6.77 + -0.52)
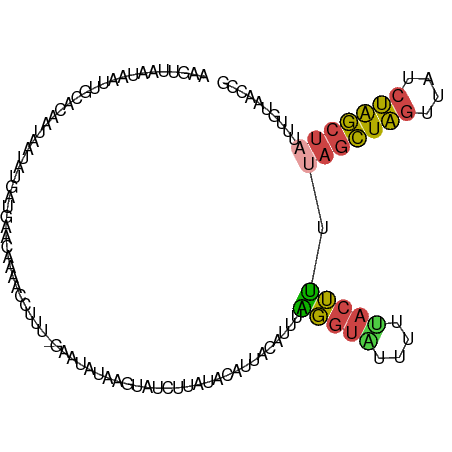
| Location | 3,199,175 – 3,199,269 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 66.35 |
| Shannon entropy | 0.61792 |
| G+C content | 0.28204 |
| Mean single sequence MFE | -15.46 |
| Consensus MFE | -6.88 |
| Energy contribution | -6.44 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
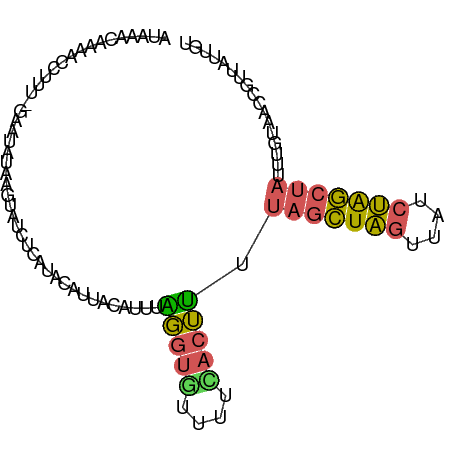
>dm3.chr3L 3199175 94 + 24543557 ACAAACAAAACCUUUUGAAUAUAAGUAUCUUAUACAUUACAUACAGGUGUUUUUACUUUUAGCUAGUUAUCUAGCUAUUUGAAACCGUUAUUGU ...(((...((((..((((((((((...))))))..)).))...))))(((((......(((((((....)))))))...))))).)))..... ( -15.30, z-score = -1.26, R) >droSim1.chr3L 2729283 93 + 22553184 AUGAACAAAACCUUU-GAAUAUAAGUAUCUCAUACAUUACAUUUAGGUGUUUUCACUUUUAGCUAGUUAUCUAGCUAUUUUUAACCGUUAUUGU ...((((((((.(((-((((.(((((((...)))).))).))))))).)))))......(((((((....))))))).........)))..... ( -15.90, z-score = -1.86, R) >droSec1.super_2 3192316 93 + 7591821 AUGAACAAAACCUUU-GACUAUAAGUAUCUCAUACAUUACAUUUAGGUGUUUUCACUUUUAGCUAGUUAUCUAGCUAUUUGUAACCGUUAUAGU ...............-.(((((((((((...)))).(((((...(((((....))))).(((((((....)))))))..)))))...))))))) ( -18.70, z-score = -2.23, R) >droEre2.scaffold_4784 5881836 94 + 25762168 AUAGAUUAAAACCUUCCAACAUAAUUAUGUUAUAACUUACAUUUGUGUACCUUAACAUUCAGCUAGUCAUCUGGCUAUUAGUAACCGUUAUUGU .................(((((....)))))(((((((((....((((......))))..((((((....))))))....))))..)))))... ( -13.10, z-score = -0.52, R) >droWil1.scaffold_180949 5025744 94 + 6375548 AUUUAAAAAACAGCCAAAAAAAAAUGAGCAAGCGCUCCCCUUGAUGAAAUCUUUACUAGUUUUUUAAUUUCGAAUCAUUUGAAAUGGCAAUUUU ............(((...(((((((((((....)))).....(((...))).......))))))).(((((((.....))))))))))...... ( -14.30, z-score = -0.51, R) >consensus AUAAACAAAACCUUU_GAAUAUAAGUAUCUCAUACAUUACAUUUAGGUGUUUUCACUUUUAGCUAGUUAUCUAGCUAUUUGUAACCGUUAUUGU ............................................(((((....))))).(((((((....)))))))................. ( -6.88 = -6.44 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:07 2011