| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,176,857 – 3,176,947 |
| Length | 90 |
| Max. P | 0.910054 |

| Location | 3,176,857 – 3,176,947 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.64654 |
| G+C content | 0.49562 |
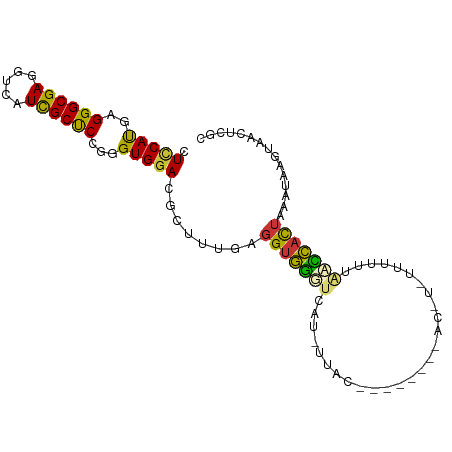
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -15.91 |
| Energy contribution | -14.80 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
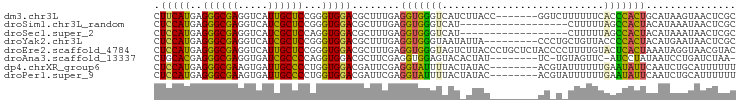
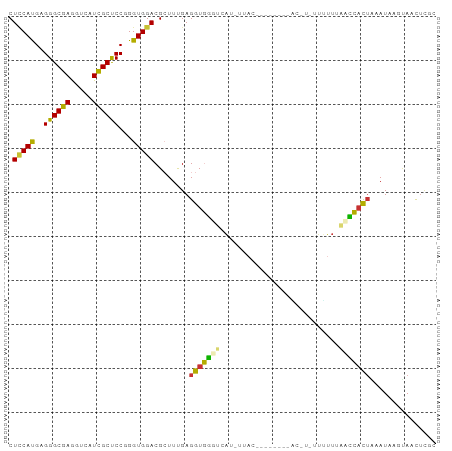
>dm3.chr3L 3176857 90 - 24543557 CUUCAUGAGGGCGAGGUCAUUGCUCCGGGUGGACGCUUUGAGGUGGGUCAUCUUACC-------GGUCUUUUUUCACCCACUGCAUAAGUAACUCGC ..........(((((.....(((...(((((((........((((((....))))))-------........)))))))...))).......))))) ( -27.99, z-score = -0.89, R) >droSim1.chr3L_random 169438 79 - 1049610 CUCCAUGAGGGCGAGGUCAUCGCUCCGGGUGGACGCUUUGAGGUGGGUCAU------------------CUUUUUAGCCACUACAUAAAUAACUCGC .(((((..((((((.....)))).))..))))).(((..((((((...)))------------------)))...)))................... ( -22.40, z-score = -0.50, R) >droSec1.super_2 3170392 79 - 7591821 CUCCAUGAGGGCGAGGUCAUCGCUCCAGGUGGACGCUUUGAGGUGGGUCAU------------------CUUUUUAGCCACUACAUAAAUAACUCGC .(((((..((((((.....)))).))..))))).(((..((((((...)))------------------)))...)))................... ( -21.40, z-score = -0.42, R) >droYak2.chr3L 3722191 88 - 24197627 CUCCAUGAGGGCGAGGUCAUCGCUCCGGGUGGACGCUUUGAGGUGGGUAAUAUUA---------CCCUGCUGUUACCCCACUACAUGAAUAACUCGC ...((((.((((((.....)))).)).(((((.((((....))))(((((((...---------......)))))))))))).)))).......... ( -26.80, z-score = -0.36, R) >droEre2.scaffold_4784 5859644 97 - 25762168 CUCCAUGAGGGCGAGGUCAUUGCUCCGGGUGGACGCUUUGAGGUGGGUAGUCUUACCCUGCUCUACCCCUUUUGUACUCACUAAAUAGGUAACGUAC .....((((..(((((........))(((((((.((...(.((((((....))))))).)))))))))...)))..))))................. ( -28.80, z-score = 0.12, R) >droAna3.scaffold_13337 14353322 86 + 23293914 CUGCACGAGGGCGAGGUGAUCGCCCCAGGUGGACGCUUCGAGGUGGAGUACACUAU--------UC-UGUAGUUC-AUCCUAUAAUCCUGAUCUAA- .......(((((((((((.(((((...))))).)))))))(((((((.((((....--------..-)))).)))-).)))....)))).......- ( -27.90, z-score = -0.90, R) >dp4.chrXR_group6 9257172 89 + 13314419 CUCCAUGAGGGCGAAGUGAUUGCCCCUGGUGGACGAUUCGAGGUAUUUUACUAUAC--------ACGUAUUUUUUGAAUAUUCAAUCUGCAUUUUUU .(((((.(((((((.....))).)))).))))).((((.(((.(((((....(((.--------...))).....)))))))))))).......... ( -20.20, z-score = -0.99, R) >droPer1.super_9 1290661 89 + 3637205 CUCCAUGAGGGCGAAGUGAUUGCCCCUGGUGGACGAUUCGAGGUAUUUUACUAUAC--------ACGUAUUUUUUGAAUAUUCAAUCUGCAUUUUUU .(((((.(((((((.....))).)))).))))).((((.(((.(((((....(((.--------...))).....)))))))))))).......... ( -20.20, z-score = -0.99, R) >consensus CUCCAUGAGGGCGAGGUCAUCGCUCCGGGUGGACGCUUUGAGGUGGGUCAU_UUAC________AC_U_UUUUUUAACCACUAAAUAAGUAACUCGC .(((((..((((((.....))))))...)))))........(((((((...........................)))))))............... (-15.91 = -14.80 + -1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:05 2011