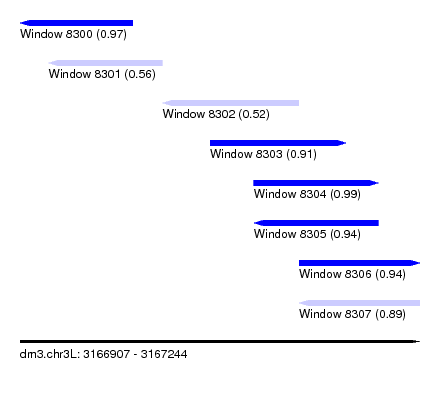
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,166,907 – 3,167,244 |
| Length | 337 |
| Max. P | 0.985682 |

| Location | 3,166,907 – 3,167,002 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.32192 |
| G+C content | 0.51188 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -23.73 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
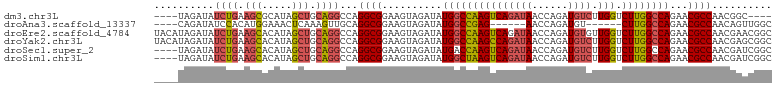
>dm3.chr3L 3166907 95 - 24543557 ----UAGAUAUCUGAAGCGCAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGGC---- ----.(((((((((.(((.....)))...(((((.((....)).....)))))............)))))))))..((((((((......))))).)))---- ( -34.20, z-score = -2.42, R) >droAna3.scaffold_13337 14343731 87 + 23293914 ----CAGAUAUCCACAUGGAAACUCAAAGUUGCAGGCGGAAGUAGAUAUGGCCGAG------AACCAGAUGU------CUUGGCCAGAACGCCAACAGUUGGC ----......(((....)))........(((....((....)).....((((((((------(........)------)))))))).)))((((.....)))) ( -24.60, z-score = -1.55, R) >droEre2.scaffold_4784 3175865 103 - 25762168 UACAUAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUGUUGGUCUUGGCCAGAACGCCAACGAACGGC .((((.(.(((((((.(((......))).(((((.((....)).....)))))...))))))).)...))))((((.(((((((......))))).)).)))) ( -34.00, z-score = -2.57, R) >droYak2.chr3L 15422585 103 + 24197627 UACAUAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGCCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGAGCGGC .....(((((((((..(((......))).(((((.((....)).....)))))............)))))))))((.(((((((......))))).)).)).. ( -35.10, z-score = -2.32, R) >droSec1.super_2 3160334 99 - 7591821 ----UAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGACCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGAUCGGC ----.......(((.(((.....))).)))(((..((....)).(((..(((((((.((..........)).)))))))(((((......)))))..)))))) ( -31.00, z-score = -1.94, R) >droSim1.chr3L 2695146 99 - 22553184 ----UAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCUAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGAUCGGC ----.(((((((((..(((......))).(((((.((....)).....)))))............)))))))))((((((((((......))))).))))).. ( -34.40, z-score = -2.52, R) >consensus ____UAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGAUCGGC ..........((((.(((.....))).))))...((((..........(((((((((((((((......)))).))).))))))))...)))).......... (-23.73 = -24.45 + 0.73)

| Location | 3,166,931 – 3,167,027 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.39521 |
| G+C content | 0.46643 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -17.59 |
| Energy contribution | -18.82 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3166931 96 - 24543557 UUCAGGGAGCCAUUGCACACAGAU----AUAGAUAUCUGAAGCGCAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUCUU .....(.(((...(((.(.(((((----(....))))))..).)))..))).).(((((.((....)).....))))).....(((((......))))). ( -27.80, z-score = -1.50, R) >droAna3.scaffold_13337 14343756 89 + 23293914 --CUGGCAUGCAACAGAUACUCGCACUGUCAGAUAUCCACAUGGAAACUCAAAGUUGCAGGCGGAAGUAGAUAUGGCCGAG------AACCAGAUGU--- --((((..((((((.((((.......)))).....(((....)))........))))))..(((..((....))..)))..------..))))....--- ( -20.60, z-score = 0.02, R) >droEre2.scaffold_4784 3175893 100 - 25762168 UUCAGGGAGCUACUGCACACAGAUAUACAUAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUGUU ....((.(((((.(((...(((((((......)))))))..)))..)))))...(((((.((....)).....)))))...........))......... ( -29.60, z-score = -2.13, R) >droYak2.chr3L 15422613 100 + 24197627 UUCAGGGAGCUGUUGCACACAGAUAUACAUAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGCCAGAUAACCAGAUGUCUU .....(.(((((((((...(((((((......)))))))..))).)))))).).(((((.((....)).....))))).....(((((......))))). ( -32.90, z-score = -2.85, R) >droSec1.super_2 3160362 96 - 7591821 UUCAGGGAGCCAUUGCACACAUAU----AUAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGACCAAGUCAGAUAACCAGAUGUCUU ........((....))........----..(((((((((..(((......))).((.((.((....)).....)).))............))))))))). ( -19.70, z-score = 0.25, R) >droSim1.chr3L 2695174 96 - 22553184 UUCAGGGAGCCAUUGCACACAGAU----AUAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCUAAGUCAGAUAACCAGAUGUCUU ........((....))....((((----((.(.(((((((.(((......))).(((((.((....)).....)))))...))))))).)...)))))). ( -24.80, z-score = -0.84, R) >consensus UUCAGGGAGCCAUUGCACACAGAU____AUAGAUAUCUGAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUCUU ........((....))..............(((((((((..(((......))).(((((.((....)).....)))))............))))))))). (-17.59 = -18.82 + 1.23)

| Location | 3,167,027 – 3,167,142 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.92 |
| Shannon entropy | 0.60319 |
| G+C content | 0.47721 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -7.71 |
| Energy contribution | -8.96 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3167027 115 - 24543557 GCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACAAUAGGCGCUCAACUUUUUGAGCUGGU---GUACAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUUCG (((((((((.......(((((.....))))))))))).(((((((((....)))))).(((---........))).)))..................))).................. ( -26.51, z-score = -2.60, R) >droAna3.scaffold_13337 14343845 93 + 23293914 ------------------GCAAAUAUUUGACAAAACAAAACACUUUUAGGCGAGAGUCUAU---GGACAAUGACCUGCCCCUCAUCCA----CUAGUCGGACCAACUUUCUCCGCUGG ------------------.......((((......)))).......(((((....)))))(---(((...(((........)))))))----((((.((((.........)))))))) ( -16.70, z-score = -0.53, R) >droEre2.scaffold_4784 3175993 112 - 25762168 GCAAUCGUUUUUACAUGUCUGACGAUCAGACGAC---AGGCGCUCAACUUUUGGACCUGGA---GUAUAAUGACCUGCCCCUCAUCCAAUUCCAAACUGCGCCAACUUUCACCAUUCG ...............((((((.....))))))..---.(((((......((((((..((((---....................))))..))))))..)))))............... ( -26.35, z-score = -2.43, R) >droYak2.chr3L 15422713 112 + 24197627 GCAAUUGUUUUUACAUGUCUGACGCUCAGACGAC---AGGCGCUCAACUUUUGGAGCUGAA---GUAUAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUUCG ...............((((((.....))))))..---.(((((......((((((...(..---(((........)))..)...))))))........)))))............... ( -24.44, z-score = -1.68, R) >droSec1.super_2 3160458 115 - 7591821 GCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACGACAGGCGCUCAACUUUUGGAGCUGGU---GUACAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUUCG .....((((......((((((.....))))))..))))(((((......((((((...((.---(((........))).))...))))))........)))))............... ( -29.74, z-score = -2.84, R) >droSim1.chr3L 2695270 115 - 22553184 GCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACGACAGGCGCUCAACUUUUGGAGCUGGU---GUACAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUUCG .....((((......((((((.....))))))..))))(((((......((((((...((.---(((........))).))...))))))........)))))............... ( -29.74, z-score = -2.84, R) >dp4.chrXR_group8 6960496 107 - 9212921 UAAAUGGAAUUGACACGAUCACUCA--AGACAAC---AAACGCCCCACCGCUAGGCCCCCUCUCAGACAGUGUCGUGUCCCUCUUU------CUGUCUCUGCCAACUUUCACCAGUCA ...((((((..(((((((.((((..--.......---....(((.........)))............))))))))))).....))------))))...................... ( -17.81, z-score = 0.28, R) >consensus GCAAUUGUCUUUACAUGUCUGAAUAUCAGACGAC___AGGCGCUCAACUUUUGGAGCUGGU___GUACAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUUCG ................(((((.....))))).......(((((((........))))............((((........))))...............)))............... ( -7.71 = -8.96 + 1.25)
| Location | 3,167,067 – 3,167,182 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Shannon entropy | 0.44684 |
| G+C content | 0.44665 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -19.03 |
| Energy contribution | -20.65 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3167067 115 + 24543557 GCAGGUCAUUGU---ACACCAGCUCAAAAAGUUGAGCGCCUAUUGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCAUUAGCAGUUGUCGUCAUACGAAUACAAUUGC ((((((......---..))).((((((....))))))((..(((((((((((.....))))).(....))))))).)).....)))....((((((((..((....))..)))))))) ( -33.60, z-score = -2.84, R) >droEre2.scaffold_4784 3176033 112 + 25762168 GCAGGUCAUUAU---ACUCCAGGUCCAAAAGUUGAGCGCCU---GUCGUCUGAUCGUCAGACAUGUAAAAACGAUUGCUCAAUUGCUUUGGCAAUUGUCGUCAUACGAAUACAAUUGC ((.((.(.....---.......).))...(((((((((...---((((((((.....)))))..((....))))))))))))))))....((((((((..((....))..)))))))) ( -29.00, z-score = -1.33, R) >droYak2.chr3L 15422753 112 - 24197627 GCAGGUCAUUAU---ACUUCAGCUCCAAAAGUUGAGCGCCU---GUCGUCUGAGCGUCAGACAUGUAAAAACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGC ((((((......---..(((((((.....))))))).))))---)).(((((.....))))).((((....(((((((.(((.....))))))))))(((.....))).))))..... ( -30.90, z-score = -1.64, R) >droSec1.super_2 3160498 115 + 7591821 GCAGGUCAUUGU---ACACCAGCUCCAAAAGUUGAGCGCCUGUCGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGC (((....(((((---(...(((((((((((((((((((..((((...(((((.....))))).......))))..)))))))))..))))).)))))(((.....))).))))))))) ( -36.60, z-score = -2.94, R) >droSim1.chr3L 2695310 115 + 22553184 GCAGGUCAUUGU---ACACCAGCUCCAAAAGUUGAGCGCCUGUCGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGC (((....(((((---(...(((((((((((((((((((..((((...(((((.....))))).......))))..)))))))))..))))).)))))(((.....))).))))))))) ( -36.60, z-score = -2.94, R) >dp4.chrXR_group8 6960530 108 + 9212921 ACACGACACUGUCUGAGAGGGGGCCUAGCGGUGGGGCGUUU---GUUGUCU--UGAGUGAUCGUGUCAAUUCCAUUUACCA--UAUCUUAUCAGCCCUUGCUCCUCCAACAGAGU--- .....((.((((.((.(((((((((....)))(((((....---((((.(.--(((....))).).))))...........--..........)))))..)))))))))))).))--- ( -28.61, z-score = 0.82, R) >consensus GCAGGUCAUUGU___ACACCAGCUCCAAAAGUUGAGCGCCU___GUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGC ((((...............(((((((((.(((((((((.........(((((.....))))).(((....)))..)))))))))...)))).)))))(((.....)))......)))) (-19.03 = -20.65 + 1.62)
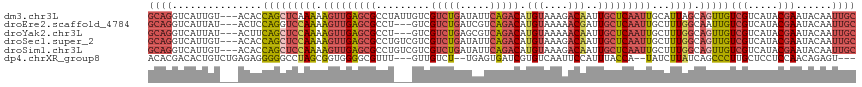
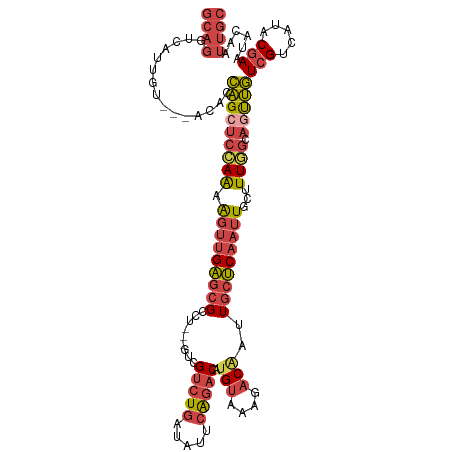
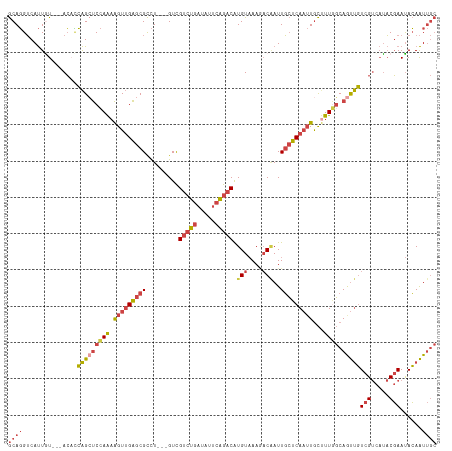
| Location | 3,167,104 – 3,167,209 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.16 |
| Shannon entropy | 0.47624 |
| G+C content | 0.40024 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3167104 105 + 24543557 UAUUGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCAUUAGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCCACCGAAA .....(((...((.(((((((...(((..(.((((((..(((((((....)))))))(((.....)))...)))))))..)))....))))))).))...))).. ( -23.60, z-score = -1.94, R) >droEre2.scaffold_4784 3176070 102 + 25762168 ---UGUCGUCUGAUCGUCAGACAUGUAAAAACGAUUGCUCAAUUGCUUUGGCAAUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCCGAAUUUCCACCGAAA ---..(((((((.....))))).........((...((......))..((((((((((..((....))..)))))))))).........))..........)).. ( -24.10, z-score = -2.14, R) >droYak2.chr3L 15422790 102 - 24197627 ---UGUCGUCUGAGCGUCAGACAUGUAAAAACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCCGAGUUUCUACCGAAA ---..(((((((.....)))))..(((.((((....((......))..((((((((((..((....))..))))))))))............)))).))).)).. ( -27.20, z-score = -2.64, R) >droSec1.super_2 3160535 105 + 7591821 UGUCGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCUACCGAAA .....(((...((.(((((((.....((((.(((((....)))))))))(((((((((..((....))..)))))))))........))))))).))...))).. ( -29.90, z-score = -3.25, R) >droSim1.chr3L 2695347 105 + 22553184 UGUCGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCUACCGAAA .....(((...((.(((((((.....((((.(((((....)))))))))(((((((((..((....))..)))))))))........))))))).))...))).. ( -29.90, z-score = -3.25, R) >dp4.chrXR_group8 6960570 91 + 9212921 ---UGUUGUCU--UGAGUGAUCGUGUCAAUUCCAUUUACCA--UAUCUUAUCAGCCCUUGCUCCUCCAACAGAGU--CCGGUCCGUACCGACAGUGGCAA----- ---.((((.(.--(((....))).).))))...........--..........(((((.((((........))))--.((((....))))..)).)))..----- ( -16.00, z-score = 0.30, R) >consensus ___UGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCCACCGAAA .......(((((.....)))))..........................((((((((((..((....))..))))))))))......................... (-15.10 = -15.97 + 0.87)
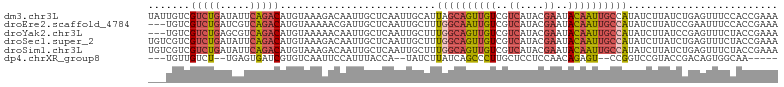
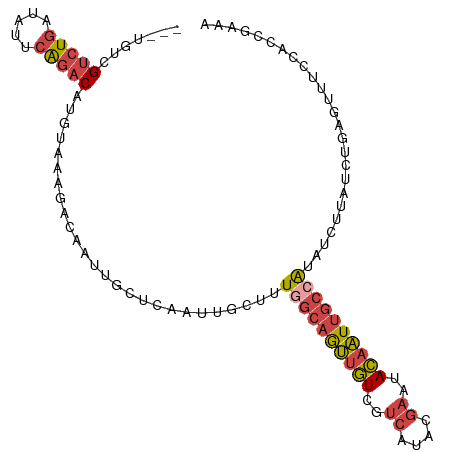
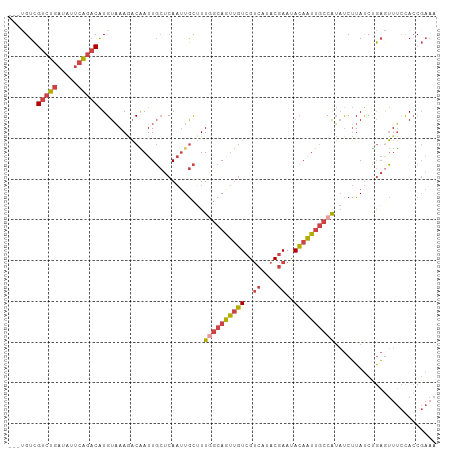
| Location | 3,167,104 – 3,167,209 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Shannon entropy | 0.47624 |
| G+C content | 0.40024 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -15.40 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.943871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3167104 105 - 24543557 UUUCGGUGGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCUAAUGCAAUUGAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACAAUA ..(((...((.(.(((((((.(....(((((((((..(((.....)))(((.(((....))).))).)))))))))....).))))))).).))...)))..... ( -28.50, z-score = -2.69, R) >droEre2.scaffold_4784 3176070 102 - 25762168 UUUCGGUGGAAAUUCGGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAAUUGCCAAAGCAAUUGAGCAAUCGUUUUUACAUGUCUGACGAUCAGACGACA--- .....((((((((...(((......((((((((((..((....))..))))))))))..((......)).))))))))))).((((((.....))))))...--- ( -28.50, z-score = -2.18, R) >droYak2.chr3L 15422790 102 + 24197627 UUUCGGUAGAAACUCGGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGAGCAAUUGUUUUUACAUGUCUGACGCUCAGACGACA--- .....((((((((............(((((.((((..((....))..)))).)))))..((......))....)))))))).((((((.....))))))...--- ( -26.90, z-score = -1.90, R) >droSec1.super_2 3160535 105 - 7591821 UUUCGGUAGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACGACA ..((((((((...((......))...(((((((((..(((.....)))(((.(((....))).))).)))))))))))))).((((((.....)))))).))).. ( -26.90, z-score = -2.16, R) >droSim1.chr3L 2695347 105 - 22553184 UUUCGGUAGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACGACA ..((((((((...((......))...(((((((((..(((.....)))(((.(((....))).))).)))))))))))))).((((((.....)))))).))).. ( -26.90, z-score = -2.16, R) >dp4.chrXR_group8 6960570 91 - 9212921 -----UUGCCACUGUCGGUACGGACCGG--ACUCUGUUGGAGGAGCAAGGGCUGAUAAGAUA--UGGUAAAUGGAAUUGACACGAUCACUCA--AGACAACA--- -----((((((.((((....(((.((..--.((((......))))...)).)))....))))--)))))).((...((((.........)))--)..))...--- ( -19.60, z-score = 0.43, R) >consensus UUUCGGUAGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACA___ ..(((...((...(((((((.(...(((((.((((..((....))..)))).)))))..((((((....)))))).....).)))))))...))...)))..... (-15.40 = -16.35 + 0.95)

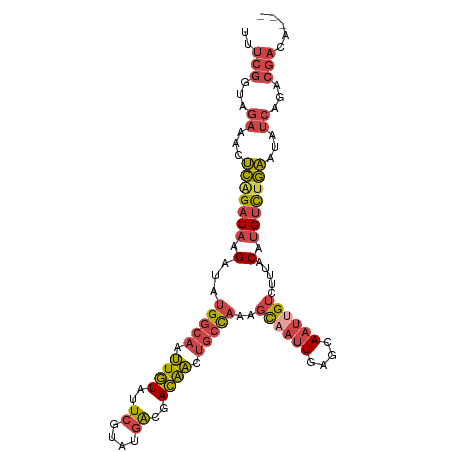
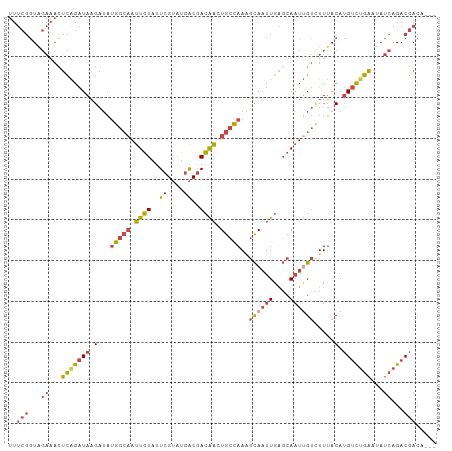
| Location | 3,167,142 – 3,167,244 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 95.87 |
| Shannon entropy | 0.07033 |
| G+C content | 0.41419 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
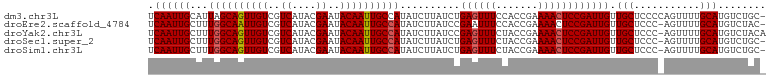
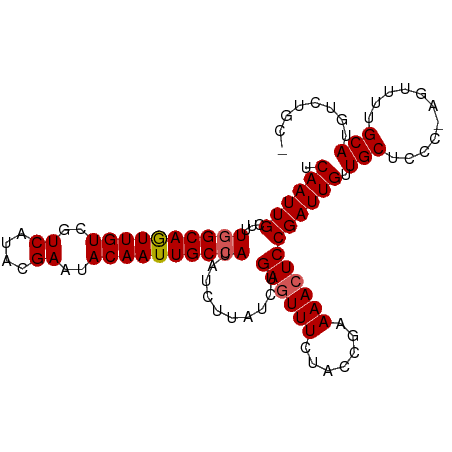
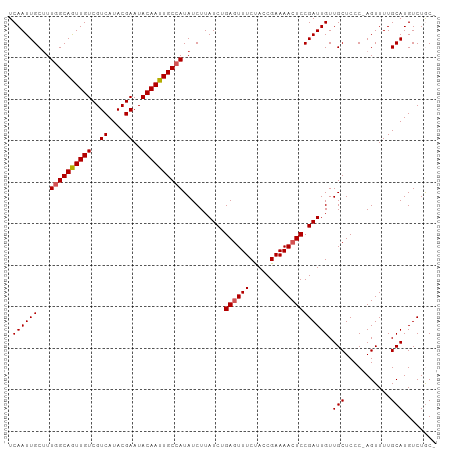
>dm3.chr3L 3167142 102 + 24543557 UCAAUUGCAUUAGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCCACCGAAAACUCCGAUUGUUGCUCCCCAGUUUUGCAUGUCUGC- .....((((...((((((((..((....))..))))))))........(((.((((((.......)))))).)))................)))).......- ( -19.10, z-score = -1.11, R) >droEre2.scaffold_4784 3176105 101 + 25762168 UCAAUUGCUUUGGCAAUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCCGAAUUUCCACCGAAAACUCCGAUUGUUGCUCCC-AGUUUUGCAUGUCUAC- .((((((...((((((((((..((....))..)))))))))).............((((....))))....)))))).(((....-......))).......- ( -19.50, z-score = -1.85, R) >droYak2.chr3L 15422825 102 - 24197627 UCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCCGAGUUUCUACCGAAAACUCCGAUUGUUGCUCCC-AGUUUUGCAUGUCUACA .((((((...((((((((((..((....))..))))))))))..........(((((((....).)))))))))))).(((....-......)))........ ( -23.40, z-score = -2.44, R) >droSec1.super_2 3160573 101 + 7591821 UCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCUACCGAAAACUCCGAUUGUUGCUCCC-AGUUUUGCAUGUCUGC- .((((((...((((((((((..((....))..))))))))))..........(((((((....).)))))))))))).(((....-......))).......- ( -23.10, z-score = -1.96, R) >droSim1.chr3L 2695385 101 + 22553184 UCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCUACCGAAAACUCCGAUUGUUGCUCCC-AGUUUUGCAUGUCUGC- .((((((...((((((((((..((....))..))))))))))..........(((((((....).)))))))))))).(((....-......))).......- ( -23.10, z-score = -1.96, R) >consensus UCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCUACCGAAAACUCCGAUUGUUGCUCCC_AGUUUUGCAUGUCUGC_ .((((((...((((((((((..((....))..))))))))))..........((((((.......)))))))))))).(((...........)))........ (-20.84 = -21.08 + 0.24)
| Location | 3,167,142 – 3,167,244 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 95.87 |
| Shannon entropy | 0.07033 |
| G+C content | 0.41419 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -23.92 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
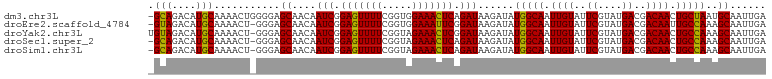
>dm3.chr3L 3167142 102 - 24543557 -GCAGACAUGCAAAACUGGGGAGCAACAAUCGGAGUUUUCGGUGGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCUAAUGCAAUUGA -(((....)))...........(((...(((.(((((((.....))))))).)))......(((((.((((..((....))..)))).))))).)))...... ( -23.80, z-score = -1.10, R) >droEre2.scaffold_4784 3176105 101 - 25762168 -GUAGACAUGCAAAACU-GGGAGCAACAAUCGGAGUUUUCGGUGGAAAUUCGGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAAUUGCCAAAGCAAUUGA -.......(((...(((-(..(((..(....)..)))..))))..................((((((((((..((....))..))))))))))..)))..... ( -26.60, z-score = -2.20, R) >droYak2.chr3L 15422825 102 + 24197627 UGUAGACAUGCAAAACU-GGGAGCAACAAUCGGAGUUUUCGGUAGAAACUCGGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGA ((((....)))).....-....((....(((.(((((((.....))))))).)))......(((((.((((..((....))..)))).)))))..))...... ( -24.30, z-score = -1.33, R) >droSec1.super_2 3160573 101 - 7591821 -GCAGACAUGCAAAACU-GGGAGCAACAAUCGGAGUUUUCGGUAGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGA -(((....)))......-....((....(((.(((((((.....))))))).)))......(((((.((((..((....))..)))).)))))..))...... ( -23.80, z-score = -1.41, R) >droSim1.chr3L 2695385 101 - 22553184 -GCAGACAUGCAAAACU-GGGAGCAACAAUCGGAGUUUUCGGUAGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGA -(((....)))......-....((....(((.(((((((.....))))))).)))......(((((.((((..((....))..)))).)))))..))...... ( -23.80, z-score = -1.41, R) >consensus _GCAGACAUGCAAAACU_GGGAGCAACAAUCGGAGUUUUCGGUAGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGA .(((....)))...........((....(((.(((((((.....))))))).)))......(((((.((((..((....))..)))).)))))..))...... (-23.92 = -23.36 + -0.56)
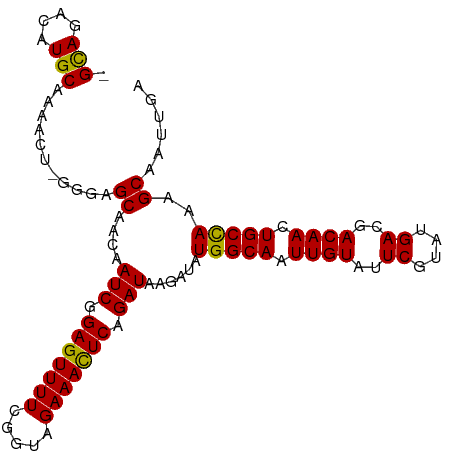
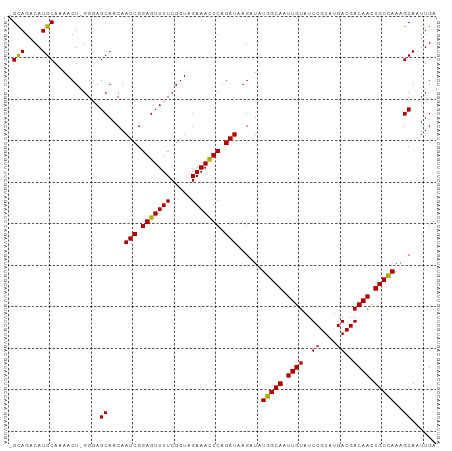
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:00:04 2011