
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,159,477 – 3,159,605 |
| Length | 128 |
| Max. P | 0.843088 |

| Location | 3,159,477 – 3,159,605 |
|---|---|
| Length | 128 |
| Sequences | 10 |
| Columns | 155 |
| Reading direction | reverse |
| Mean pairwise identity | 61.88 |
| Shannon entropy | 0.73627 |
| G+C content | 0.35458 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -7.17 |
| Energy contribution | -5.93 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.76 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
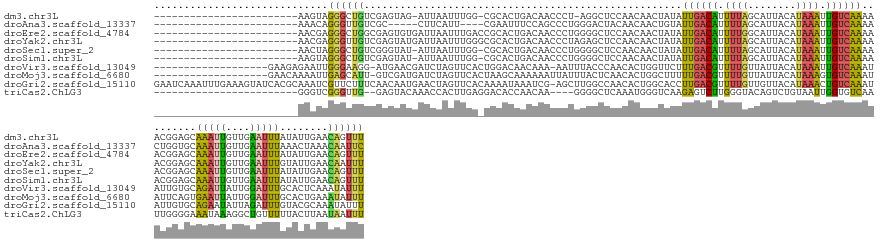
>dm3.chr3L 3159477 128 - 24543557 ------------------------AAGUAGGGCUGUCGAGUAG-AUUAAUUUGG-CGCACUGACAACCCU-AGGCUCCAACAACUAUAUUGACAUUUUAGCAUUACAUAAAUUGUCAAAAACGGAGCAAAUUGUUGAAUUUAUAUUGAACAGUUU ------------------------...(((((.(((((.((.(-(.....)).)-)....))))).))))-).(((((..........((((((.((((........)))).))))))....)))))((((((((.(((....))).)))))))) ( -32.04, z-score = -2.62, R) >droAna3.scaffold_13337 14336969 122 + 23293914 ------------------------AAACAGGGUUGUCGC-----CUUCAUU----CGAAUUUCCAGCCCUGGGACUACAACAACUGUAUUGACAUUUUAGCAUUACAUAAAUUGUCAAAACUGGUGCAAAUUGUUGAAUUUAAACUAAACAAUUC ------------------------...((((((((....-----.(((...----.)))....))))))))......((((((.((((((((((.((((........)))).)))))......)))))..))))))................... ( -27.20, z-score = -1.95, R) >droEre2.scaffold_4784 3168553 131 - 25762168 ------------------------AACGAGGGCUGGCGAGUGUGAUUAAUUUGACCGCACUGACAACCCUGGGGCUCCAACAACUAUAUUGACAUUUUGGCAUUACAUAAAUUGUCAAAAACGGAGCAAAUUGUUGAAUUUAUAUUGAACAGUUU ------------------------..(.((((.((...((((((.((.....)).))))))..)).)))).).(((((..(((.....)))...((((((((..........))))))))..)))))((((((((.(((....))).)))))))) ( -37.90, z-score = -3.39, R) >droYak2.chr3L 15415219 131 + 24197627 ------------------------AACGAGGGUUGUCGAGUAUGAUUAAUUUGGGCGCACUGACAACCCUAGAGCUCCAACAACUAUAUUGACAUUUUAGCAUUACAUAAAUUGUCAAAAACGGAGCAAAUUGUUGAAUUUGUAUUGAACAAUUU ------------------------....(((((((((.(((.((...........)).))))))))))))...(((((..........((((((.((((........)))).))))))....)))))((((((((.(((....))).)))))))) ( -36.14, z-score = -3.70, R) >droSec1.super_2 3153139 129 - 7591821 ------------------------AACUAGGGCUGUCGGGUAU-AUUAAUUUGG-CGCACUGACAACCCUGGGGCUCCAACAACUAUAUUGACAUUUUAGCAUUACAUAAAUUGUCAAAAACGGAGCAAAUUGUUGAAUUUAUAUUGAACAGUUU ------------------------..((((((.((((((((.(-(......)).-.)).)))))).)))))).(((((..........((((((.((((........)))).))))))....)))))((((((((.(((....))).)))))))) ( -39.14, z-score = -4.52, R) >droSim1.chr3L 2687796 129 - 22553184 ------------------------AAGUAGGGCUGUCGAGUAU-AUUAAUUUGG-CGCACUGACAACCCUGGGGCUCCAACAACUAUAUUGACAUUUUAGCAUUACAUAAAUUGUCAAAAACGGAGCAAAUUGUUGAAUUUAUAUUGAACAGUUU ------------------------...(((((.((((.(((..-..........-...))))))).)))))..(((((..........((((((.((((........)))).))))))....)))))((((((((.(((....))).)))))))) ( -31.60, z-score = -2.03, R) >droVir3.scaffold_13049 7550965 134 - 25233164 -------------------GAAGAGAAUUGGGAAGG-AUGAACGAUCUAGUUCACUGGACAACAAA-AAUUUACCCAACACUGGUUCUUUGACGUUUUGUUAUUACAUAAAUUGUCAAAUAUUGUGCAGAUUAUUGGAUUUGCACUCAAAUAUUU -------------------..(((((((..((..((-.(((((......))))).((.....))..-.......))..).)..)))))))((((.(((((......))))).))))((((((((((((((((....)))))))))...))))))) ( -32.10, z-score = -2.26, R) >droMoj3.scaffold_6680 5803592 135 - 24764193 -------------------GAACAAAAUUGAGCAUU-GUCGAUGAUCUAGUUCACUAAGCAAAAAAUUAUUUACUCAACACUGGCUUUUUGACGUUUUGUUAUUACAUAAAGUGUCAAAUAUUCAGUGAAUUAUUGGAUUUGCACUGAAAUAUUU -------------------............(((..-.((((((((.(((....))).....................((((((...(((((((((((((......)))))))))))))...)))))).))))))))...)))............ ( -26.90, z-score = -1.20, R) >droGri2.scaffold_15110 19236846 154 + 24565398 GAAUCAAAUUUGAAAGUAUCACGCAAAUCGUUCUUUCAACAAUGAACUAGUUCACAAAAUAAAUCG-AGCUUGGCCAACACUGGCACCUUGACGUUUUGUUGUUACAUAAACUGUCAAAUAUUGUGCAGAAUAUUAGAUUUGUACGCAAAUAUUU .......(((((...((.....(((((((.....(((.((((((..(.(((((............)-)))).)((((....))))...((((((((((((....))).)))).))))).))))))...))).....))))))))).))))).... ( -30.30, z-score = -0.75, R) >triCas2.ChLG3 10761413 125 - 32080666 ------------------------GGGUCGGGUUG--GAGUACAAACCACUUGAGGACACCAACAA----GGGGCUCAAAUGGGUCAAGAGUCUUGGGUACAGUCUGUAAUUGGUGUCAAUUGGGGAAAUAAAGGCUGUUUUUACUUAAUAAUUU ------------------------.((((((.(((--.....))).)).((..(.(((((((((((----(((((((....))))).....)))))..(((.....))).))))))))..)..))........)))).................. ( -31.10, z-score = -0.44, R) >consensus ________________________AAAUAGGGCUGUCGAGUAUGAUUAAUUUGA_CGCACUGACAACCCUGGGGCUCCAACAACUAUAUUGACAUUUUAGCAUUACAUAAAUUGUCAAAAACGGAGCAAAUUGUUGAAUUUAUACUGAACAAUUU .............................((((((.....................................................((((((.((((........)))).)))))).........(((((....)))))........)))))) ( -7.17 = -5.93 + -1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:57 2011