| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,139,427 – 3,139,527 |
| Length | 100 |
| Max. P | 0.717975 |

| Location | 3,139,427 – 3,139,527 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.26 |
| Shannon entropy | 0.56084 |
| G+C content | 0.43184 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -13.52 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.717975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3139427 100 - 24543557 UACAUGUACAUAUCUCUGGUUUUUCUUCGCUUCUUCGUUUUGGCGCAGGAAAAACCAAAUGUGGCGGACAAUUUUCCCCUUGGAAACCGAAUUUCCUUCA----------------- ........(((((...(((((((((((((((..........)))).))))))))))).))))).(((.....(((((....))))))))...........----------------- ( -25.60, z-score = -2.13, R) >droAna3.scaffold_13337 14316738 78 + 23293914 ----------UAUGUGGGUAUUUUUCUGGCUGCCAUGCUUUGGUGGAGGGGGA-------GCGGAGGAUGAUUUCCCUUUCAAGGAACAUCGUCC---------------------- ----------..........(((..((..(..(((.....)))..).))..))-------)....(((((((((((.......)))).)))))))---------------------- ( -22.90, z-score = -0.06, R) >droEre2.scaffold_4784 3148506 93 - 25762168 ------UAUGUAUCUCUGGUUUUUCUUCGCUUCUUCGUUUUGGCACAGGAAAAACCAAAUGUGGCGGAGAAUUUUCCCCUUGGAAAGCGAAUUUCCUCA------------------ ------..........(((((((((((.(((..........)))..))))))))))).....((.((((....))))))..(((((.....)))))...------------------ ( -22.70, z-score = -0.81, R) >droYak2.chr3L 15394228 93 + 24197627 ------UAUGUAUCUCUGCUUUUUCUUCGCAUCUUCGUUUUGGCGCAGGAAAAACCAAAUGUGGCGGAGAAUUUUCCCCUUGAAAACCGAAUUUUCUCA------------------ ------......((((((((((((((((((............))).))))))).........))))))))...........(((((.....)))))...------------------ ( -18.40, z-score = -0.21, R) >droSec1.super_2 3133082 117 - 7591821 UAUAUGUACAUGUCUCUGGAUUUUCGUCGCUUCUUCGUUUUGGCGCAGGGAAAACCAAAUGUGGCGAACAAUUUUCCCCUUGGAAAAUUUCUUCCUUUGGAAACCGAAUUUCCUUCA .................(((..((((..........((((.(.((((.((....))...)))).)))))...(((((....((((......))))...))))).))))..))).... ( -25.40, z-score = -0.19, R) >droSim1.chr3L 2667104 100 - 22553184 UAUAUGUACAUGUCUCUGGUUUUUCUUCGCUUCUUCGUUUUGGCGCAGGAAAAACCAAAUGUGGCGGACAAUUUUCCCCUUGGAAACCGAAUUUCCUUCA----------------- .......((((.....(((((((((((((((..........)))).))))))))))).))))((.(((......)))))..(((((.....)))))....----------------- ( -26.30, z-score = -2.08, R) >consensus ______UACAUAUCUCUGGUUUUUCUUCGCUUCUUCGUUUUGGCGCAGGAAAAACCAAAUGUGGCGGACAAUUUUCCCCUUGGAAACCGAAUUUCCUUA__________________ ................(((((((((((((((..........)))).))))))))))).....((.(((......)))))..(((((.....)))))..................... (-13.52 = -14.33 + 0.81)

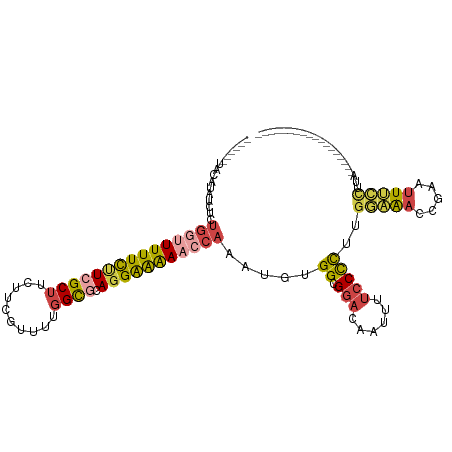
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:53 2011