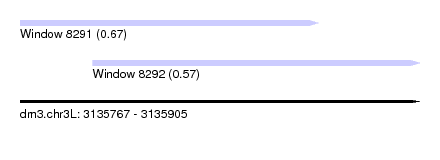
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,135,767 – 3,135,905 |
| Length | 138 |
| Max. P | 0.671870 |

| Location | 3,135,767 – 3,135,870 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.03 |
| Shannon entropy | 0.23554 |
| G+C content | 0.45681 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -24.82 |
| Energy contribution | -27.10 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
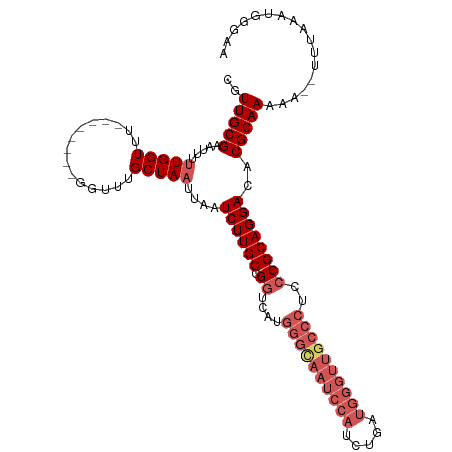
>dm3.chr3L 3135767 103 + 24543557 CGUUGCGAAUUUUUGGUUU--------GGUUUGCUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUCCGAUGGGUUGCCCUCCCGCAGGACACGCAAAAA--UUCAAAUGGGAA ((((..(((((((((((((--------((....))))....((((((.((....(((((((((((...))))))))))).)).)))))).)).))))))--))).)))).... ( -39.70, z-score = -3.11, R) >droEre2.scaffold_4784 3144389 89 + 25762168 CGUUGCGAAUUUUUGGUUU--------GGUUUGCUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUC----------------CGCAGGACACGCAAAAAAUUUUAAAUGGCCA ..(((((.....(((((..--------.....)))))....((((((.((..((((.....)))))----------------)))))))..)))))................. ( -19.60, z-score = 0.59, R) >droYak2.chr3L 15390348 110 - 24197627 CGUUGCGAAUUUCUGGUUUUCUGGUUUGGUUUGCUAAUUAAUCUUGCCGGUCAUGGGUAAUCCAUCUGAUGGGUUGCCC--CCGCAGGACACGCAAAAAAUUU-AAAUGGCCA ..(((((...(((((.....(((((..((((........))))..)))))....(((((((((((...)))))))))))--...)))))..))))).......-......... ( -33.90, z-score = -1.36, R) >droSec1.super_2 3129262 102 + 7591821 CGUUGCGAAUUUUUGGUUU--------GGUUUGCUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUCUGAUGGGUUGCCCUCCCGCAGGACACGCAAAA---UUUAAAUGGGAA ..(((((.....(((((..--------.....)))))....((((((.((....(((((((((((...))))))))))).)).))))))..)))))..---............ ( -36.10, z-score = -2.55, R) >droSim1.chr3L 2663284 103 + 22553184 CGUUGCGAAUUUUUGGUUU--------GGUUUGCUAAUUAAUCUUGCCGGCCAUGGGCAAUCCAUCUGAUGGGUUGCCCUCCCGCAGGACACGCAAAAA--UUUAAAUGGGAA ((((..(((((((((((((--------((....))))....((((((.((....(((((((((((...))))))))))).)).)))))).)).))))))--))).)))).... ( -37.20, z-score = -2.44, R) >consensus CGUUGCGAAUUUUUGGUUU________GGUUUGCUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUCUGAUGGGUUGCCCUCCCGCAGGACACGCAAAAA__UUUAAAUGGGAA ..(((((.....(((((...............)))))....((((((.((....((((((((((.....))))))))))..))))))))..)))))................. (-24.82 = -27.10 + 2.28)

| Location | 3,135,792 – 3,135,905 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.09 |
| Shannon entropy | 0.20850 |
| G+C content | 0.51296 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -30.50 |
| Energy contribution | -31.86 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
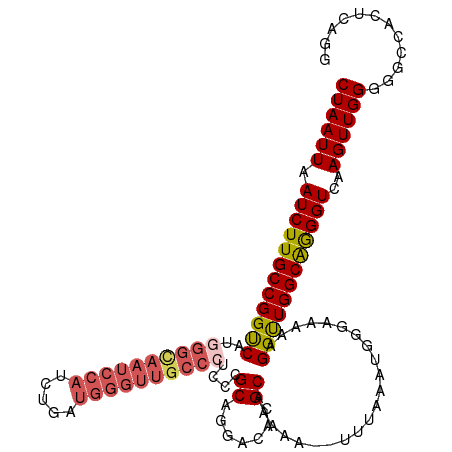
>dm3.chr3L 3135792 113 + 24543557 CUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUCCGAUGGGUUGCCCUCCCGCAGGACACGCAAAAA--UUCAAAUGGGAAAAAGGUUGGCAGGGUCAAGUUGGGGGCCACUCAGG ((((((.(((((((((..(..(((((((((((...)))))))))))(((((...((..........--.))...)))))....)..)))))))))..))))))............ ( -41.50, z-score = -1.86, R) >droEre2.scaffold_4784 3144414 99 + 25762168 CUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUC----------------CGCAGGACACGCAAAAAAUUUUAAAUGGCCAAAAGGUUGGCAAGGUCAAGUUGGGGGCCACUCAGG ........((((((.((..((((.....)))))----------------)))))))..................(((((...((.(((((...))))).))...)))))...... ( -25.80, z-score = -0.33, R) >droYak2.chr3L 15390381 112 - 24197627 CUAAUUAAUCUUGCCGGUCAUGGGUAAUCCAUCUGAUGGGUUGCC--CCCGCAGGACACGCAAAAAAUUU-AAAUGGCCAAAUGGCUGGCGAGGUCAAGUUGGGGGCCACUCAGG ((((((.(((((((((((((((((((((((((...))))))))))--)..((.......)).........-..........))))))))))))))..))))))............ ( -40.70, z-score = -1.69, R) >droSec1.super_2 3129287 112 + 7591821 CUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUCUGAUGGGUUGCCCUCCCGCAGGACACGCAAAA---UUUAAAUGGGAAAAAGGUUGGCAGGGUCAAGUUGGGGGCCACUCAGG ((((((.(((((((((..(..(((((((((((...)))))))))))((((((.......))...(---(....))))))....)..)))))))))..))))))............ ( -40.10, z-score = -1.65, R) >droSim1.chr3L 2663309 113 + 22553184 CUAAUUAAUCUUGCCGGCCAUGGGCAAUCCAUCUGAUGGGUUGCCCUCCCGCAGGACACGCAAAAA--UUUAAAUGGGAAAAAGGUUGGCAGGGUCAAGUUGGGGGCCACUCAGU ((((((.((((((((((((..(((((((((((...)))))))))))((((((.......))....(--(....))))))....))))))))))))..))))))............ ( -44.30, z-score = -2.43, R) >consensus CUAAUUAAUCUUGCCGGUCAUGGGCAAUCCAUCUGAUGGGUUGCCCUCCCGCAGGACACGCAAAAA__UUUAAAUGGGAAAAAGGUUGGCAGGGUCAAGUUGGGGGCCACUCAGG ((((((.((((((((((((..((((((((((.....))))))))))....((.......))......................))))))))))))..))))))............ (-30.50 = -31.86 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:51 2011