| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,097,508 – 3,097,598 |
| Length | 90 |
| Max. P | 0.996230 |

| Location | 3,097,508 – 3,097,598 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.95 |
| Shannon entropy | 0.34241 |
| G+C content | 0.50082 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -30.81 |
| Energy contribution | -30.15 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
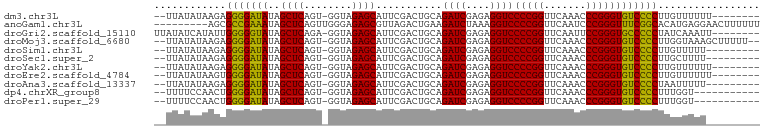
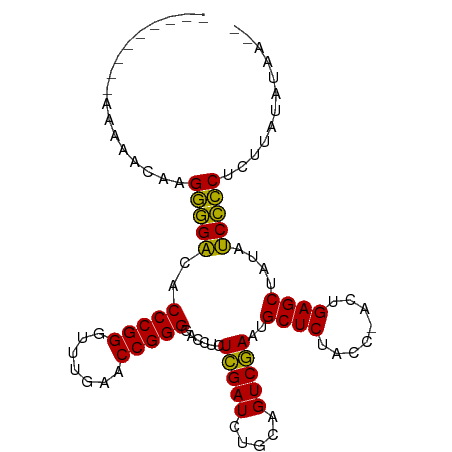
>dm3.chr3L 3097508 90 + 24543557 --UUAUAUAAGAGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUUUU-------- --........(((((((((..((((...-....))))...........((((....))))(((((.......)))))))))))))).......-------- ( -32.50, z-score = -2.70, R) >anoGam1.chr3L 37406798 92 + 41284009 ---------AGCGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCACAUGAGGAACUUUUUU ---------.(.(((((((..((((........))))...........((((....))))(((((.......)))))))))))).)............... ( -27.30, z-score = -0.51, R) >droGri2.scaffold_15110 15870285 92 - 24565398 UUAUAUCAUAUUGGGGGUAUAGCUCAGA-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAUUCCGGGUGCCCCCUAUCAAAUU-------- ............(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))).........-------- ( -32.30, z-score = -2.27, R) >droMoj3.scaffold_6680 23768423 96 - 24764193 --UUAUAUAAGAGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAAGCUUUUU-- --((((....(((((((((..((((...-....))))...........((((....))))(((((.......)))))))))))))).))))........-- ( -33.40, z-score = -2.18, R) >droSim1.chr3L 2622090 89 + 22553184 --UUAUAUAAGAGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUUU--------- --........(((((((((..((((...-....))))...........((((....))))(((((.......))))))))))))))......--------- ( -32.50, z-score = -2.73, R) >droSec1.super_2 3091196 89 + 7591821 --UUAUAUAAGAGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGCUUUU--------- --........(((((((((..((((...-....))))...........((((....))))(((((.......))))))))))))))......--------- ( -32.50, z-score = -2.41, R) >droYak2.chr3L 15348565 90 - 24197627 --UUAUAUAAGAGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUUUU-------- --........(((((((((..((((...-....))))...........((((....))))(((((.......)))))))))))))).......-------- ( -32.50, z-score = -2.70, R) >droEre2.scaffold_4784 3105867 90 + 25762168 --UUAUAUAAGUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUUUU-------- --..........(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))).........-------- ( -29.80, z-score = -1.87, R) >droAna3.scaffold_13337 14272371 89 - 23293914 --UUAUAUAAGAGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAAUUUUU--------- --.........((((((((..((((...-....))))...........((((....))))(((((.......))))))))))))).......--------- ( -31.30, z-score = -2.57, R) >dp4.chrXR_group8 3962612 87 + 9212921 --UUUUCCAACUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUUGGU----------- --....((((..(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))).)))).----------- ( -33.50, z-score = -2.06, R) >droPer1.super_29 194155 87 + 1099123 --UUUUCCAACUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUUGGU----------- --....((((..(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))).)))).----------- ( -33.50, z-score = -2.06, R) >consensus __UUAUAUAAGAGGGGAUAUAGCUCAGU_GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUUU_________ ............(((((((..((((........))))...........((((....))))(((((.......))))))))))))................. (-30.81 = -30.15 + -0.66)
| Location | 3,097,508 – 3,097,598 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.95 |
| Shannon entropy | 0.34241 |
| G+C content | 0.50082 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -29.44 |
| Energy contribution | -28.78 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.996230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
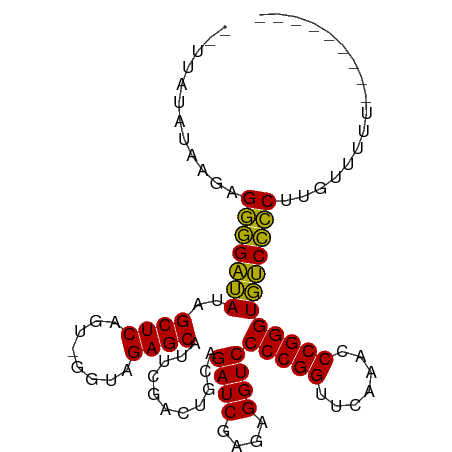
>dm3.chr3L 3097508 90 - 24543557 --------AAAAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCUCUUAUAUAA-- --------........((((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))).........-- ( -29.00, z-score = -2.63, R) >anoGam1.chr3L 37406798 92 - 41284009 AAAAAAGUUCCUCAUGUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCGCU--------- ...............((((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))).--------- ( -33.00, z-score = -3.60, R) >droGri2.scaffold_15110 15870285 92 + 24565398 --------AAUUUGAUAGGGGGCACCCGGAAUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-UCUGAGCUAUACCCCCAAUAUGAUAUAA --------.........(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))............ ( -29.10, z-score = -1.96, R) >droMoj3.scaffold_6680 23768423 96 + 24764193 --AAAAAGCUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCUCUUAUAUAA-- --..............((((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))).........-- ( -29.00, z-score = -1.98, R) >droSim1.chr3L 2622090 89 - 22553184 ---------AAAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCUCUUAUAUAA-- ---------.......((((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))).........-- ( -29.00, z-score = -2.58, R) >droSec1.super_2 3091196 89 - 7591821 ---------AAAAGCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCUCUUAUAUAA-- ---------..(((..((((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))))))......-- ( -29.30, z-score = -2.21, R) >droYak2.chr3L 15348565 90 + 24197627 --------AAAAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCUCUUAUAUAA-- --------........((((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))).........-- ( -29.00, z-score = -2.63, R) >droEre2.scaffold_4784 3105867 90 - 25762168 --------AAAAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCACUUAUAUAA-- --------.........(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..........-- ( -28.90, z-score = -2.74, R) >droAna3.scaffold_13337 14272371 89 + 23293914 ---------AAAAAUUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCUCUUAUAUAA-- ---------.......((((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))).........-- ( -28.90, z-score = -2.47, R) >dp4.chrXR_group8 3962612 87 - 9212921 -----------ACCAAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAGUUGGAAAA-- -----------.((((.(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..))))....-- ( -32.50, z-score = -2.27, R) >droPer1.super_29 194155 87 - 1099123 -----------ACCAAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAGUUGGAAAA-- -----------.((((.(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..))))....-- ( -32.50, z-score = -2.27, R) >consensus _________AAAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC_ACUGAGCUAUAUCCCCUCUUAUAUAA__ .................(((((..(((((.......)))))......(((((.....)))))..((((........))))....)))))............ (-29.44 = -28.78 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:46 2011