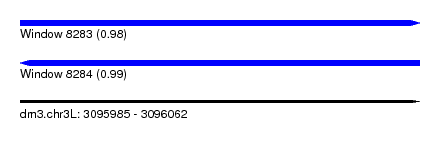
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,095,985 – 3,096,062 |
| Length | 77 |
| Max. P | 0.992154 |

| Location | 3,095,985 – 3,096,062 |
|---|---|
| Length | 77 |
| Sequences | 13 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 93.82 |
| Shannon entropy | 0.14961 |
| G+C content | 0.56989 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -30.28 |
| Energy contribution | -29.85 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 1.02 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
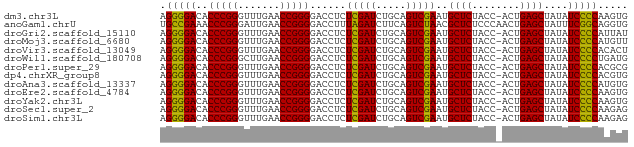
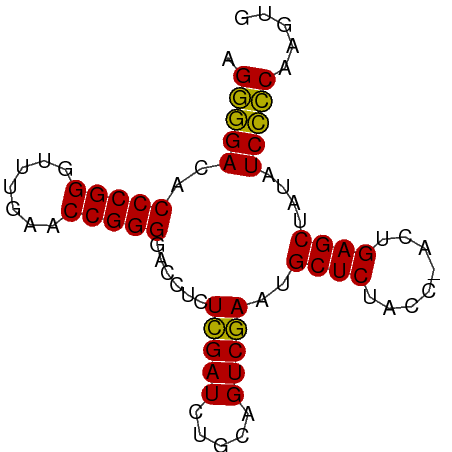
>dm3.chr3L 3095985 77 + 24543557 CACUUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.80, R) >anoGam1.chrU 31108598 78 + 59568033 CACCUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCA .....(((((((..((((........))))...........((((....))))(((((.......)))))))))))). ( -26.70, z-score = -1.50, R) >droGri2.scaffold_15110 1480101 77 + 24565398 AUAAUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -2.21, R) >droMoj3.scaffold_6680 23767399 77 - 24764193 AACAUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -2.07, R) >droVir3.scaffold_13049 145074 77 + 25233164 AGUGUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.90, R) >droWil1.scaffold_180708 1890682 77 + 12563649 CAUCAGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAGCCCGGGUGUCCCCU ....((((((((..((((...-....))))...........((((....))))(((((.......))))))))))))) ( -30.50, z-score = -2.14, R) >droPer1.super_29 192737 77 + 1099123 CGCGUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.57, R) >dp4.chrXR_group8 3961190 77 + 9212921 CACGUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.77, R) >droAna3.scaffold_13337 14271275 77 - 23293914 CACAUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.94, R) >droEre2.scaffold_4784 3104153 77 + 25762168 CACUUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.80, R) >droYak2.chr3L 15346018 77 - 24197627 CACUUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.80, R) >droSec1.super_2 3089507 77 + 7591821 CUCUUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.62, R) >droSim1.chr3L 2618867 77 + 22553184 CUCUUGGGGAUAUAGCUCAGU-GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((...-....))))...........((((....))))(((((.......)))))))))))). ( -29.80, z-score = -1.62, R) >consensus CACUUGGGGAUAUAGCUCAGU_GGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCU .....(((((((..((((........))))...........((((....))))(((((.......)))))))))))). (-30.28 = -29.85 + -0.42)

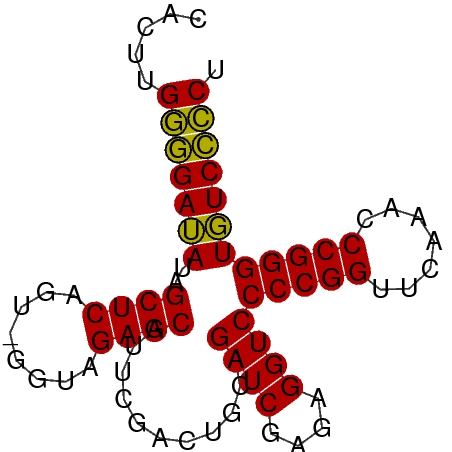
| Location | 3,095,985 – 3,096,062 |
|---|---|
| Length | 77 |
| Sequences | 13 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Shannon entropy | 0.14961 |
| G+C content | 0.56989 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -29.45 |
| Energy contribution | -29.02 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.19 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3095985 77 - 24543557 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAAGUG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -1.96, R) >anoGam1.chrU 31108598 78 - 59568033 UGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAGGUG (((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))).... ( -30.30, z-score = -3.24, R) >droGri2.scaffold_15110 1480101 77 - 24565398 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAUUAU .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -2.44, R) >droMoj3.scaffold_6680 23767399 77 + 24764193 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAUGUU .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -2.03, R) >droVir3.scaffold_13049 145074 77 - 25233164 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCACACU .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -2.42, R) >droWil1.scaffold_180708 1890682 77 - 12563649 AGGGGACACCCGGGCUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCUGAUG ((((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))).... ( -29.30, z-score = -2.45, R) >droPer1.super_29 192737 77 - 1099123 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCACGCG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -2.12, R) >dp4.chrXR_group8 3961190 77 - 9212921 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCACGUG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -1.99, R) >droAna3.scaffold_13337 14271275 77 + 23293914 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAUGUG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -1.99, R) >droEre2.scaffold_4784 3104153 77 - 25762168 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAAGUG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -1.96, R) >droYak2.chr3L 15346018 77 + 24197627 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAAGUG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -1.96, R) >droSec1.super_2 3089507 77 - 7591821 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAAGAG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -1.98, R) >droSim1.chr3L 2618867 77 - 22553184 AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC-ACUGAGCUAUAUCCCCAAGAG .(((((..(((((.......)))))......(((((.....)))))..((((....-...))))....)))))..... ( -28.90, z-score = -1.98, R) >consensus AGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACC_ACUGAGCUAUAUCCCCAAGUG .(((((..(((((.......)))))......(((((.....)))))..((((........))))....)))))..... (-29.45 = -29.02 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:45 2011