
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,095,717 – 3,095,869 |
| Length | 152 |
| Max. P | 0.977201 |

| Location | 3,095,717 – 3,095,869 |
|---|---|
| Length | 152 |
| Sequences | 11 |
| Columns | 156 |
| Reading direction | forward |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.56910 |
| G+C content | 0.47959 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
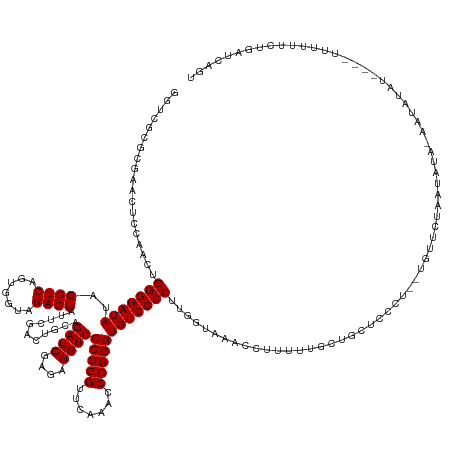
>dm3.chr3L 3095717 152 + 24543557 GGUCACUCGAAUCCCAACUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACCUUUUUGUUGCUCCCCCCAGUCCGAAUACAAAGUAUAU----UUUUUUCUGAUCACU (((((......((...(((((((....(((.(((.(((...(((......)))..((((((.((.((((((.......))))).).)).))))))..)))...)))..)))..)))))))..))((((...))))..----.......)))))... ( -44.20, z-score = -0.95, R) >droSim1.chr3L 2618613 150 + 22553184 GGUCGCGCGAACUCCAACUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACCUUUUUGCUGCUCCCC--AGUUCAAAUAUAAAGUAUAU----UUUUUUCUGAUCAGU ((((...........(((((((((..((((.....(((...(((......)))..((((((.((.((((((.......))))).).)).))))))..))).....)))).)))))--)))).(((((((...)))))----))......))))... ( -47.90, z-score = -1.95, R) >droSec1.super_2 3089240 149 + 7591821 GGUCGCGCGAACUCCAACUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACCUUUUUGCUGCUCCC---AGUUCAAAUAUAAAGUAUAU----UUUUUUCUGAUCAGC ((((...........((((((((...((((.....(((...(((......)))..((((((.((.((((((.......))))).).)).))))))..))).....)))).))))---)))).(((((((...)))))----))......))))... ( -43.60, z-score = -0.83, R) >droYak2.chr3L 15345744 156 - 24197627 GGUCGCGCGAACUCCAACUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACCUUUUUGUUGCUGCCUGCUCCCCAGUUCUAAAAUGUAUAUAAUUUUCUCUGAUCAGU (((((.(.((.....(((((((((....((.((((..(((((((......)))..((((((.((.((((((.......))))).).)).)))))).......))))..))))...)))))))))))...((((.......)))))).))))))... ( -48.00, z-score = -1.26, R) >droEre2.scaffold_4784 3103888 149 + 25762168 GGUCGCGCGGAGUGCAACUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACCUUUUUGCUCCCC-----AGUUUUACUAAAAUUUAUAUAU--UUUUGUCUGAUCAGU (((((.(((.((((.(((((((((....((((.......))))........((((((((((.((.((((((.......))))).).)).))))))........)))))))))-----)))).))))(((((......))--)))))).)))))... ( -50.20, z-score = -2.29, R) >droAna3.scaffold_13337 14271035 143 - 23293914 AGCCGCGCUUGCUCCAACUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACUUUUUUGUUCUUUU------UCCAACCUU--UCUUUAU-----UUUUCCUGAUCAAU .((.(((..(((((..((((((.......))))))....)))))..)).).)).(((((.((((.((((((.......))))).).))..((((..(((......))).....------.))))....--.......-----...)).)))))... ( -39.10, z-score = -0.71, R) >dp4.chrXR_group8 3960930 155 + 9212921 GGCCGGUCGAAGCCCAUCUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGAGUAACCUUUUUUCCUCUUUUUUUUUUACUUAACUC-AACAUUUCAGCUUUCUUUUGAUCAAU .((.(((((((.(((....)))......((((.......)))).))))))))).((((((((((..(((((.......)))))(((....((((((((....................))))))))...-.))).........))))))))))... ( -44.35, z-score = -1.30, R) >droPer1.super_29 192478 154 + 1099123 GGCCGGUCGAAGCCCAUCUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGAGUAACCUUUUUUCCUCUUUUUUUUU-ACUUAUCUC-AACAUUUCAGCGUUCUUUUGAUCAAU .((.(((((((.(((....)))......((((.......)))).))))))))).((((((((((..(((((.......)))))(((.....(((((((...................))-)))))....-.))).........))))))))))... ( -44.01, z-score = -1.09, R) >droVir3.scaffold_13049 144849 116 + 25233164 AGUCGCUCGGUUUGCAUCUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAGGUAAAC-UUUUUAUUAUAUACAAC--------------------------------------- ........(((((((....(((((((..((((.......))))...........((((....))))(((((.......))))))))))))...))))))-)................--------------------------------------- ( -37.20, z-score = -1.60, R) >droMoj3.scaffold_6680 23767141 133 - 24764193 AGUCGCUGGCAACGUAUCUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAGGUAAACUUUUUUCACAUUAUUUGUUUAUUUAAAAUG-AAUG---------------------- .......(....).(((((((((.....((((.......))))....(((....((((....))))(((((.......))))).))))))))))))..............((((..(((.....)))..-))))---------------------- ( -37.10, z-score = -0.85, R) >droGri2.scaffold_15110 1479851 131 + 24565398 AGUCGUCGCAGUUGUAUCUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAGGUAAACUUUUUUGUACACAAUGCUUUUUUUAGAAUA-AG------------------------ .......((((((.(((((((((.....((((.......))))....(((....((((....))))(((((.......))))).)))))))))))))))....(((....)))))).............-..------------------------ ( -37.90, z-score = -1.26, R) >consensus GGUCGCGCGAACUCCAACUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACCUUUUUGCUGCUCCCU__UGUUCUAAUAUA_AAUAUAU____UUUUUUCUGAUCAGU ...................(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).................................................................. (-29.80 = -29.80 + 0.00)

| Location | 3,095,717 – 3,095,869 |
|---|---|
| Length | 152 |
| Sequences | 11 |
| Columns | 156 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.56910 |
| G+C content | 0.47959 |
| Mean single sequence MFE | -42.37 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
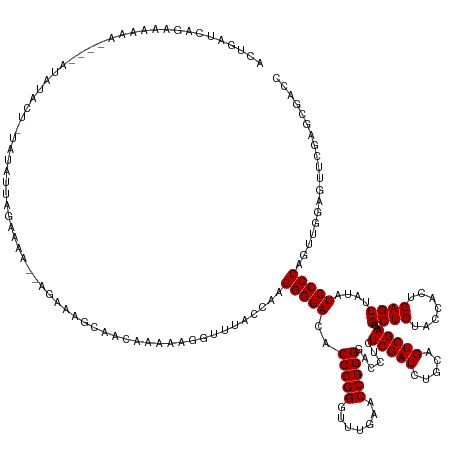
>dm3.chr3L 3095717 152 - 24543557 AGUGAUCAGAAAAAA----AUAUACUUUGUAUUCGGACUGGGGGGAGCAACAAAAAGGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGUUGGGAUUCGAGUGACC ...............----...(((((...((((.(((((((((.(((........((((((((..(((....))).)))..)))))(((((.(..(((((.....)))))..).))).)).....)))...))))))))).))))..)))))... ( -50.70, z-score = -1.43, R) >droSim1.chr3L 2618613 150 - 22553184 ACUGAUCAGAAAAAA----AUAUACUUUAUAUUUGAACU--GGGGAGCAGCAAAAAGGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGUUGGAGUUCGCGCGACC ...............----...........((((.((((--(((((((((......((....)).(((((...(((((.......)))))..)))))....))))........((((.......))))....))))))))).)))).......... ( -47.70, z-score = -1.88, R) >droSec1.super_2 3089240 149 - 7591821 GCUGAUCAGAAAAAA----AUAUACUUUAUAUUUGAACU---GGGAGCAGCAAAAAGGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGUUGGAGUUCGCGCGACC ((((.((((....((----(((((...)))))))...))---))...)))).....((((......(((....)))(..(((.(((.(((((....(((((.....)))))..((((.......))))....))))).))).)))..)....)))) ( -46.40, z-score = -1.71, R) >droYak2.chr3L 15345744 156 + 24197627 ACUGAUCAGAGAAAAUUAUAUACAUUUUAGAACUGGGGAGCAGGCAGCAACAAAAAGGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGUUGGAGUUCGCGCGACC ..............................(((((((((((((((...........((....)).(((((...(((((.......)))))..))))).).)))))........((((.......))))....)))))))))............... ( -46.50, z-score = -0.86, R) >droEre2.scaffold_4784 3103888 149 - 25762168 ACUGAUCAGACAAAA--AUAUAUAAAUUUUAGUAAAACU-----GGGGAGCAAAAAGGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGUUGCACUCCGCGCGACC ...............--.............(((..((((-----((((((((....((....)).(((((...(((((.......)))))..))))).....)))........((((.......))))....)))))))))..))).......... ( -44.40, z-score = -1.68, R) >droAna3.scaffold_13337 14271035 143 + 23293914 AUUGAUCAGGAAAA-----AUAAAGA--AAGGUUGGA------AAAAGAACAAAAAAGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGUUGGAGCAAGCGCGGCU ...((((.......-----.......--..(((....------....((((......))))))).(((((...(((((.......)))))..))))).))))...(((((..(((((((..((((...........)))))))))))....))))) ( -39.50, z-score = -0.38, R) >dp4.chrXR_group8 3960930 155 - 9212921 AUUGAUCAAAAGAAAGCUGAAAUGUU-GAGUUAAGUAAAAAAAAAAGAGGAAAAAAGGUUACUCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGAUGGGCUUCGACCGGCC ...((((.......((((.(.....)-.)))).............(((((................(....).(((((.......)))))..))))).)))).((.((((((.((((.......))))......(((....))).))))))..)). ( -41.60, z-score = -0.30, R) >droPer1.super_29 192478 154 - 1099123 AUUGAUCAAAAGAACGCUGAAAUGUU-GAGAUAAGU-AAAAAAAAAGAGGAAAAAAGGUUACUCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGAUGGGCUUCGACCGGCC ...............((((...(((.-.((((....-.........(((............))).(((((...(((((.......)))))..)))))..)))))))((((((.((((.......))))......(((....))).)))))))))). ( -42.00, z-score = -0.45, R) >droVir3.scaffold_13049 144849 116 - 25233164 ---------------------------------------GUUGUAUAUAAUAAAAA-GUUUACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGAUGCAAACCGAGCGACU ---------------------------------------(((((............-.........(((....))).((((((..(.(((((....(((((.....)))))..((((.......))))....))))).)...))))))..))))). ( -36.10, z-score = -2.22, R) >droMoj3.scaffold_6680 23767141 133 + 24764193 ----------------------CAUU-CAUUUUAAAUAAACAAAUAAUGUGAAAAAAGUUUACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGAUACGUUGCCAGCGACU ----------------------..((-(((.(((..........))).)))))....((((((((.(((....)))))))..)))).(((((....(((((.....)))))..((((.......))))....)))))......((((....)))). ( -33.90, z-score = -1.19, R) >droGri2.scaffold_15110 1479851 131 - 24565398 ------------------------CU-UAUUCUAAAAAAAGCAUUGUGUACAAAAAAGUUUACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGAUACAACUGCGACGACU ------------------------..-.............(((((((((........((((((((.(((....)))))))..)))).(((((....(((((.....)))))..((((.......))))....)))))..)))))).)))....... ( -37.30, z-score = -2.41, R) >consensus ACUGAUCAGAAAAAA____AUAUACU_UAUAUUAGAAAA__AGAAAGCAACAAAAAGGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAGUUGGAGUUCGAGCGACC ..................................................................(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))................... (-28.90 = -28.90 + 0.00)
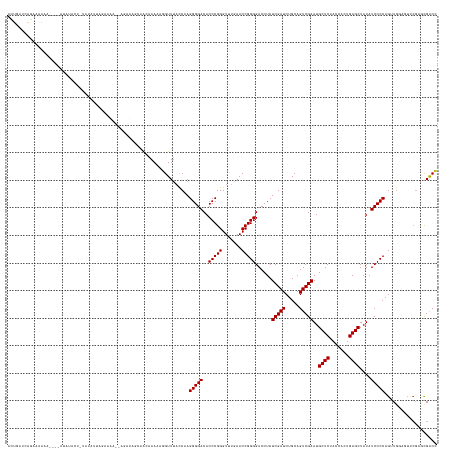
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:43 2011