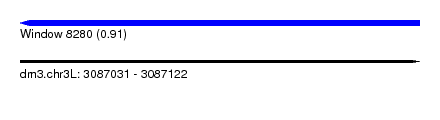
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,087,031 – 3,087,122 |
| Length | 91 |
| Max. P | 0.913805 |

| Location | 3,087,031 – 3,087,122 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 65.93 |
| Shannon entropy | 0.75847 |
| G+C content | 0.44449 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -10.34 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3087031 91 - 24543557 UAAAUAAUUAAUAAUUUGAGCAACAAUAUGAUCAUGGUC-AAAGAAAAACGAACCUGAAGGGGCGGGCCUCGCCUUGGAAUGGAUCAGUGUA ......................(((...(((((....((-((.......((..((....))..))(((...)))))))....))))).))). ( -18.10, z-score = -0.34, R) >droGri2.scaffold_15110 1469591 85 - 24565398 -------CAAUUAAAUGUGGGGCAUGCCUCAUUGAGGUUUGUCGCCUCACUCACCUGAAGGGGCGUGCGUCGCCUUGGAAUGGAUCGGUGUA -------((.((.((.((((.(((((((((..((((((.....)))))).((....)).))))))))).)))).)).)).)).......... ( -29.50, z-score = -1.10, R) >droVir3.scaffold_13049 133914 87 - 25233164 -GAUUAAGUGUUAGAUUUAAUGGGAAUUAGAAUUCGAU---UUUUG-AACUCACCUGAAGGGGCGUGCCUCGCCUUGGAAUGGAUCUGUGUA -..........((((((((.((((..((((((......---)))))-).))))((....((((((.....))))))))..)))))))).... ( -17.70, z-score = 0.43, R) >droSim1.chr3L 2609881 91 - 22553184 CAAAUAAUUAAUAGUUGGAGCAACAAUAUGAUCAUGGUC-UAAGAAAAACGAACCUGAAGGGGCGGGCCUCGCCUUGGAAUGGAUCAGUGUA ......................(((...(((((....((-(((......((..((....))..))(((...))))))))...))))).))). ( -19.60, z-score = -0.35, R) >droSec1.super_2 3080491 91 - 7591821 CAAAUAAUUAAUAGUUGGAGCAACAAUAUGAUCAUGGUC-UACGAAAAACGAACCUGAAGGGGCGGGCCUCGCCUUGGAAUGGAUCAGUGUA ......................(((...(((((....((-((.......((..((....))..))(((...))).))))...))))).))). ( -18.50, z-score = 0.24, R) >droYak2.chr3L 15337081 91 + 24197627 CAAUUAAUUAAUAGUUGGUGGAACAACAUGAUUAUGAUC-AAUAAAAAACGCACCUGAAGGGGCGGGCCUCGCCUUGGAAUGGAUCAGUGUA ....((((((...((((......)))).))))))(((((-.........(((.((.....)))))(((...)))........)))))..... ( -19.20, z-score = -0.10, R) >droEre2.scaffold_4784 3095158 91 - 25762168 CAAAUAAUCAAUAGUUGGAUCAACUACAUGAUCAUGGUC-AGAGAAAAACGCACCUGAAGGGGCGGGCCUCGCCUUGGAAUGGAUCAGUGUA ............(((((...)))))((((((((....((-((.......(((.((.....)))))(((...)))))))....))))).))). ( -20.00, z-score = 0.33, R) >droAna3.scaffold_13337 14262411 89 + 23293914 AAAAAUGAUAACAUUUUAGAGAAGAAUUAAAAACCAGU---UAAGGAAACUCACCUGAAGGGGCGGGCAUCGCCUUGGAAUGGAUCAGAGUA .....((((..(((((((................(((.---..((....))...)))...(((((.....)))))))))))).))))..... ( -18.10, z-score = -0.86, R) >dp4.chrXR_group8 3949345 80 - 9212921 ----------UCAAAUCAUCUGCAUUGAUCCGUUUGGUU--UUGGUUUACCCACCUGAAUGGGCGAGCUUCGCCUUGGAAUGGAUCAGUGUA ----------..........((((((((((((((((((.--..((((..((((......))))..))))..)))...))))))))))))))) ( -32.10, z-score = -4.16, R) >droPer1.super_29 180707 80 - 1099123 ----------UCAACUCAUCUGCAUUGAUCCGUUUGGUU--CUGGUUUACCCACCUGAAUGGGCGAGCUUCGCCUUGGAAUGGAUCAGUGUA ----------..........((((((((((((((((((.--..((((..((((......))))..))))..)))...))))))))))))))) ( -32.10, z-score = -3.98, R) >droMoj3.scaffold_6680 23756821 87 + 24764193 UAAACGAACAUUAGUUACGAUAUAAGUAUACAGGCUAU-----AGCAGACUCACCUGAAGGGGCGUGCCUCGCCUUGGAAUGGAUCUGUGUA ....(((((....))).)).......((((((((((((-----..(((......)))..((((((.....))))))...)))).)))))))) ( -21.40, z-score = -0.41, R) >consensus _AAAUAAUUAAUAGUUGGAGCAACAAUAUGAUUAUGGUC__AAGAAAAACGCACCUGAAGGGGCGGGCCUCGCCUUGGAAUGGAUCAGUGUA .....................................................((....((((((.....)))))).....))......... (-10.34 = -10.52 + 0.18)

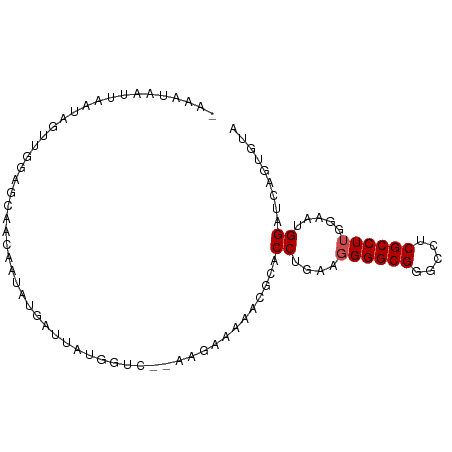
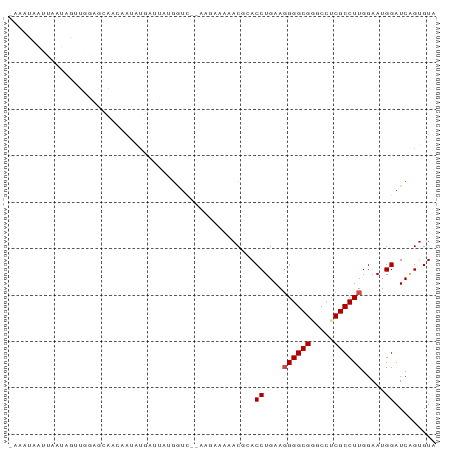
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:41 2011