| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,083,837 – 3,083,902 |
| Length | 65 |
| Max. P | 0.998804 |

| Location | 3,083,837 – 3,083,902 |
|---|---|
| Length | 65 |
| Sequences | 7 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 56.19 |
| Shannon entropy | 0.90722 |
| G+C content | 0.45033 |
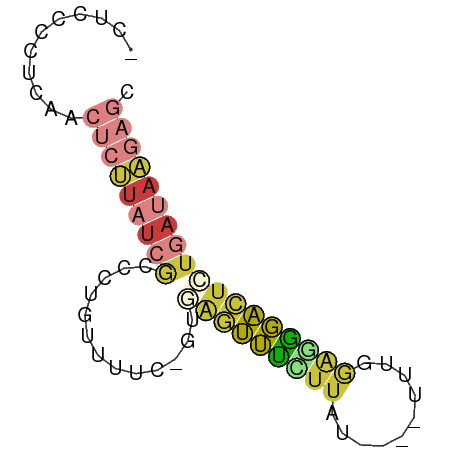
| Mean single sequence MFE | -15.67 |
| Consensus MFE | -7.89 |
| Energy contribution | -8.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.72 |
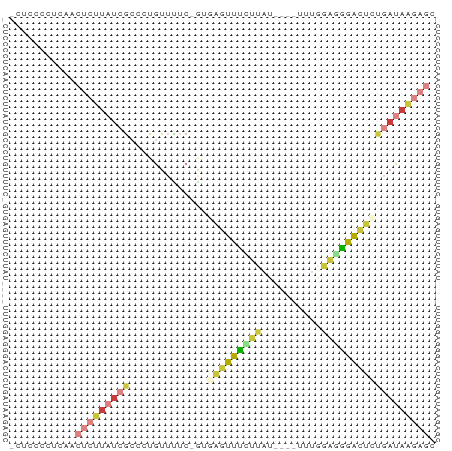
| Mean z-score | -1.59 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.998804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
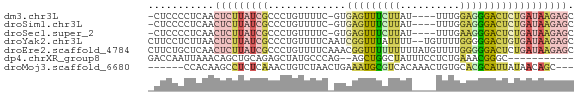
>dm3.chr3L 3083837 65 - 24543557 -CUCCCCUCAACUCUUAUCGCCCUGUUUUC-GUGAGUUUCUUAU----UUUGGAGGGACUCUGAUAAGAGC -..........((((((((.....(....)-..((((..(((..----....)))..)))).)))))))). ( -17.90, z-score = -1.92, R) >droSim1.chr3L 2606730 65 - 22553184 -CUCCCCUCAACUCUUAUCGCCCUGUUUUC-GUGAGUUUCUUAU----UUUGGAGGGACUCUGAUAAGAGC -..........((((((((.....(....)-..((((..(((..----....)))..)))).)))))))). ( -17.90, z-score = -1.92, R) >droSec1.super_2 3077350 65 - 7591821 -CUCCCCUCAACUCUUAUCGCCCUGUUUUC-GUGAGUUUCUUAU----UUUGAAGGGACUCUGAUAAGAGC -..........((((((((.....(....)-..((((..(((..----....)))..)))).)))))))). ( -17.50, z-score = -2.44, R) >droYak2.chr3L 15333930 69 + 24197627 CUUCCUCUUAACUCUUAUCGCCCUGUUUUCAAUCGGUUUAUUUU--UGUUUUGGGGGACUGUGAUAAGAGC ...........((((((((((...(((..(...(((.......)--)).....)..))).)))))))))). ( -17.70, z-score = -2.40, R) >droEre2.scaffold_4784 3092022 71 - 25762168 CUUCUGCUCAACUCUUAUCGCCCUGUUUUCAAACGGUUUUUUUUUAUGUUUUGGGGGACUCUGAUAAGAGC ...........(((((((((....(((..(.((((...........))))...)..)))..))))))))). ( -15.10, z-score = -1.23, R) >dp4.chrXR_group8 3946224 58 - 9212921 GACCAAUUAAACAGCUGCAGAGCUAUGCCCAG--AGCUGGCUAUUUCCUCUGAAACGGGC----------- ..((.......((((((((......)))....--)))))....((((....)))).))..----------- ( -10.80, z-score = 0.95, R) >droMoj3.scaffold_6680 23752738 62 + 24764193 ------CCACAAGCCUCUCAAACUGUCUAACUGAAAUGCGUCACAAACUGUGCACGCAUUAUAACAGC--- ------................((((.....((.((((((((((.....))).))))))).)))))).--- ( -12.80, z-score = -2.21, R) >consensus _CUCCCCUCAACUCUUAUCGCCCUGUUUUC_GUGAGUUUCUUAU____UUUGGAGGGACUCUGAUAAGAGC ...........(((((((((.............(((((((((..........)))))))))))))))))). ( -7.89 = -8.90 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:39 2011